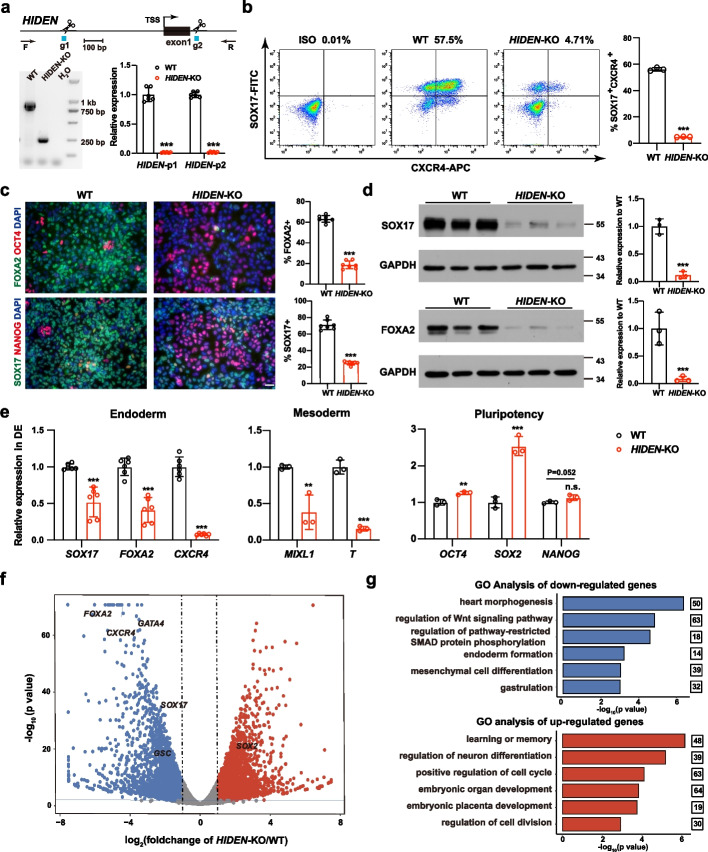
Fig. 2.
HIDEN is a desert lncRNA required for endoderm differentiation. a Generation of HIDEN knockout (KO) PSCs by CRISPR/Cas9. Top: sgRNAs used to delete the promoter of HIDEN and genomic PCR primers used to detect the promoter deletion. Bottom: The HIDEN RNA level was quantified using two sets of primers in HIDEN-KO DE cells compared to wildtype (n = 6). b Flow cytometric analysis of SOX17+CXCR4+ cells in wildtype and HIDEN-KO DE cells. The statistical results were shown on the right (n = 3). c Immunofluorescent staining of DE markers (SOX17, FOXA2) and pluripotency markers (OCT4, NANOG) in wildtype and HIDEN-KO DE cells. Quantitative results were shown on the right (n = 7). Scale bar, 50 m. d The protein levels of DE markers (SOX17 and FOXA2) were determined in wildtype and HIDEN-KO DE cells (n = 3). GAPDH was used as internal control. e The RNA expression levels of representative endoderm genes (n = 6), mesoderm genes (n = 3) and pluripotency genes (n = 3) in wildtype and HIDEN-KO DE cells. f Scatterplot showing differentially expressed genes identified by RNA-seq of wildtype and HIDEN-KO DE cells (n = 3). Upregulated and downregulated genes upon HIDEN-KO were shown in red and blue, respectively. g GO analysis of the downregulated or upregulated genes in HIDEN-KO DE cells compared to wildtype