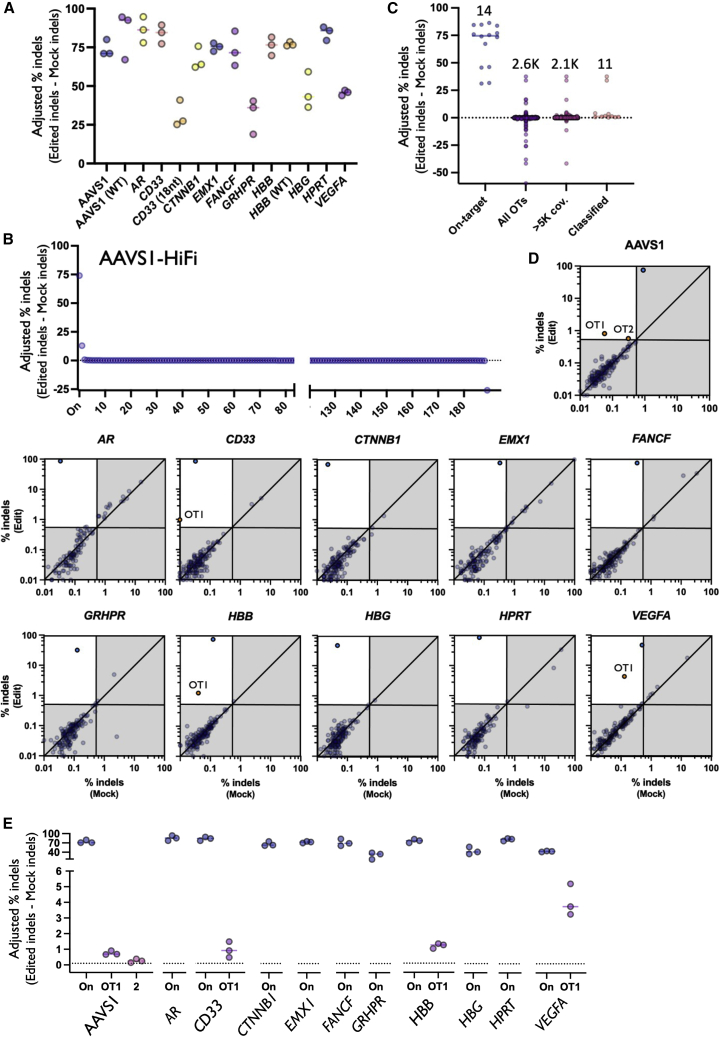
Figure 2.
Summary of on-target and OT editing
(A) On-target activity of each sgRNA determined by inclusion in rhAmpSeq NGS panel. All treatments were performed using HiFi Cas9 with 20-nt gRNA unless otherwise noted in parentheses. N = 3 separate HSPC donors per treatment. (B) Each dot depicts percent adjusted indels (Edit-Mock) for each donor at the on-target as well as each OT site in each category. Numbers above dots indicate the total number of sites in each category. (C) Each dot depicts the average percent indels (Edit-Mock) across donors at each site on panel in AAVS1-HiFi treatment (including the on-target site). The dotted line depicts 0.1% adjusted indel detection threshold after Mock is subtracted from Edited treatments. All sites are shown before filtering. (D) Each dot depicts the percent indels averaged across donors for each site on panel with average coverage or more than 5,000×. Top left quadrant indicates indels more than 0.5% indels in Edit treatment and less than 0.4% indels in Mock treatment. Blue dots represent on-target indels and orange dots represent classified OT sites. Shown on base 10 logarithmic scale. (E) Each dot depicts the percent indels (Edit-Mock) for each donor at the on-target as well as each OT site that remained after filtering using HiFi Cas9 and 20-nt spacers. The solid bars depict the median percent indels for all three HSPC donors. The dotted line depicts 0.1% adjusted indel detection threshold after Mock is subtracted from Edited treatments.