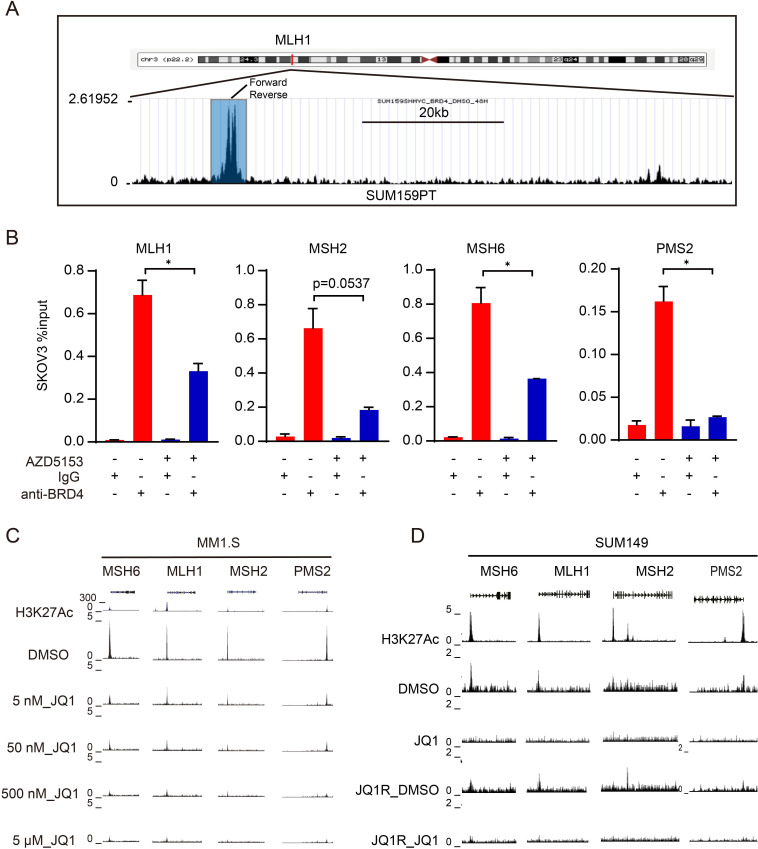
Figure 3.
BRD4 transcriptionally regulates mismatch repair genes (A) UCSC Genome Browser was used to show BRD4 ChIP sequencing signal profiles in the MLH1 gene locus. Primers for ChIP-qPCR validation were indicated. (B) ChIP-qPCR of BRD4 in SKOV3 treated with vehicle or 1 µM AZD5135 for 48 hours. Data is shown as mean±SEM from each of three independent replicates. Two-tailed t-tests: *, p<0.05; **, p<0.01; ***, p<0.001. (C) ChIP sequencing showed BRD4 binding at the MLH1, MSH2, MSH6, and PMS2 loci in MM1.S cells treated with DMSO or the indicated dose of JQ1 in GSE44931 data set. (D) ChIP sequencing showed BRD4 binding at the MLH1, MSH2, MSH6, and PMS2 loci in SUM149 and JQ1-resistant (JQ1R) SUM149 cells treated with DMSO or 10 µM JQ1 in GSE131102 data set. BRD4, bromodomain containing 4; ChIP, chromatin immunoprecipitation; MLH1, mutL homologue 1; MSH2, mutS homologue 2; MSH6, mutS homologue 6; PMS2, PMS1 homolog 2, mismatch repair system component; qPCR, quantitative PCR; DMSO, Dimethyl sulfoxide.