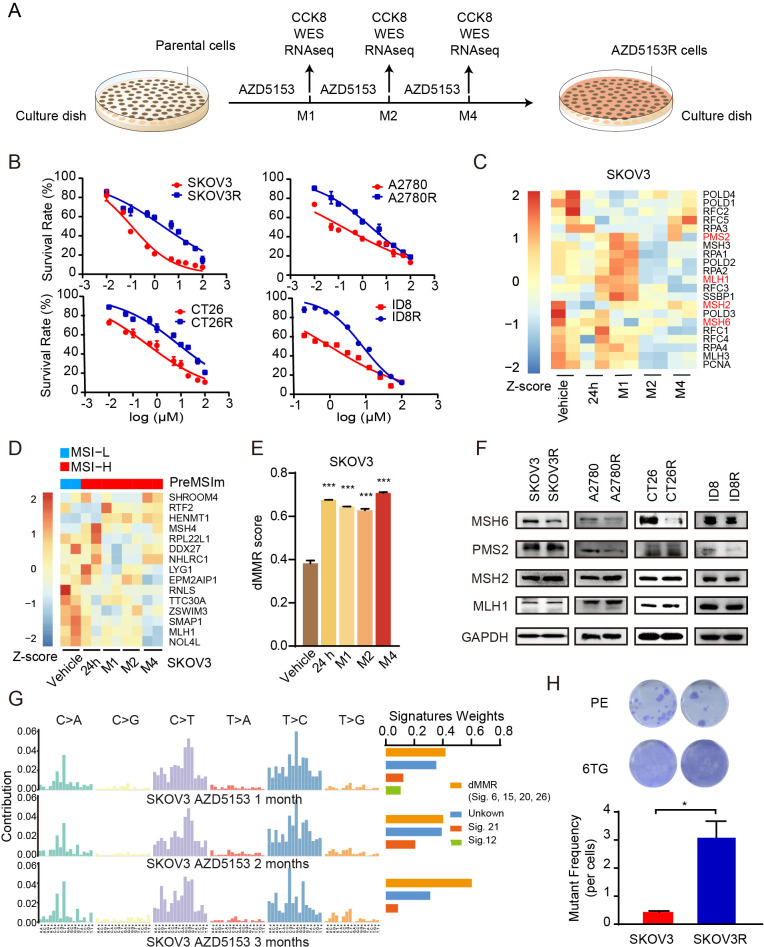
Figure 5.
Prolonged incubation with BRD4i in vitro results in acquisition of dMMR, a dMMR mutational signature that persists despite BRD4i resistance (A) Schematic diagram of the establishment of AZD5153-resistant cell lines. Parental cells were subjected to gradual increases in AZD5153 concentrations until cells grew in the presence of 15–20-fold half-maximal inhibitory concentration (2–4 months from initial exposure). Samples were collected at 24 hours, 1 month, 2 month, and 4 month for RNA-seq and WES. AZD5153-induced cells with massive resistance after 2–4-month exposure were recognized as AZD5153R cell lines. (B) Cell viability curves of parental or AZD5153-resistant (A2780R (M2), SKOV3R (M4), CT26R (M4), and ID8R (M4)) cells treated with AZD5153 for 48 hours. Representative results are presented as means±SEM of three independent experiments. (C) RNA-seq data from SKOV3 cells treated with vehicle or AZD5153 (24 hours, 1 month, 2 months, and 4 months) were analyzed for the expression of 23 MMR-related genes. Heatmap shows z-score of relative expression of 23 MMR-related genes in indicated treatment. (D) RNA-seq data from SKOV3 cells treated with vehicle or AZD5153 (24 hours, 1 month, 2 months, and 4 months) were analyzed for PreMSIm signature to assess the MSI status. Heatmap shows z-score of relative expression of genes in indicated time point of AZD5153 treatment. The top bar indicates the MSI status defined by PreMSIm (blue indicates MSI-low (MSI-L), representing MMR-proficient; red indicates MSI-high (MSI-H), representing dMMR). (E) RNA-seq data from SKOV3 cells treated with vehicle or AZD5153 (24 hours, 1 month, 2 months, and 4 months) were analyzed for dMMR scores. Data represent mean±SD, two-tailed t-tests: *, p<0.05; **, p<0.01; ***, p<0.001. (F) Western blotting of MLH1, MSH2, MSH6, and PMS2 expression levels in A2780R (M2), SKOV3R (M4), and CT26R (M4) cells and their parental cells. (G) WES data from SKOV3 cells treated with vehicle or AZD5153 (1 month, 2 months, and 4 months) were analyzed for the mutational signature and the weights of known mutational processes in the COSMIC Signatures for each sample. The proportion of dMMR mutation signatures represented the sum of weights for dMMR-related COSMIC Signatures at each time point. (H) Representative pictures (upper) and mutant frequency (lower) of mutability assay in parental SKOV3 cells and SKOV3R (M4) cells. Data is shown as mean±SEM from each of three independent replicates, two-tailed t-tests: *, p<0.05; **, p<0.01; ***, p<0.001. BRD4i, bromodomain containing 4 inhibitors; dMMR, MMR deficiency; MLH1, mutL homologue 1; MMR, mismatch repair; MSH2, mutS homologue 2; MSH6, mutS homologue 6; MSI, microsatellite instability; PE, plating efficiency; PMS2, PMS1 homolog 2, mismatch repair system component; RNA-seq, RNA sequencing; WES, Whole Exome Sequencing; 6-TG, 6-thioguanine.