Figure.
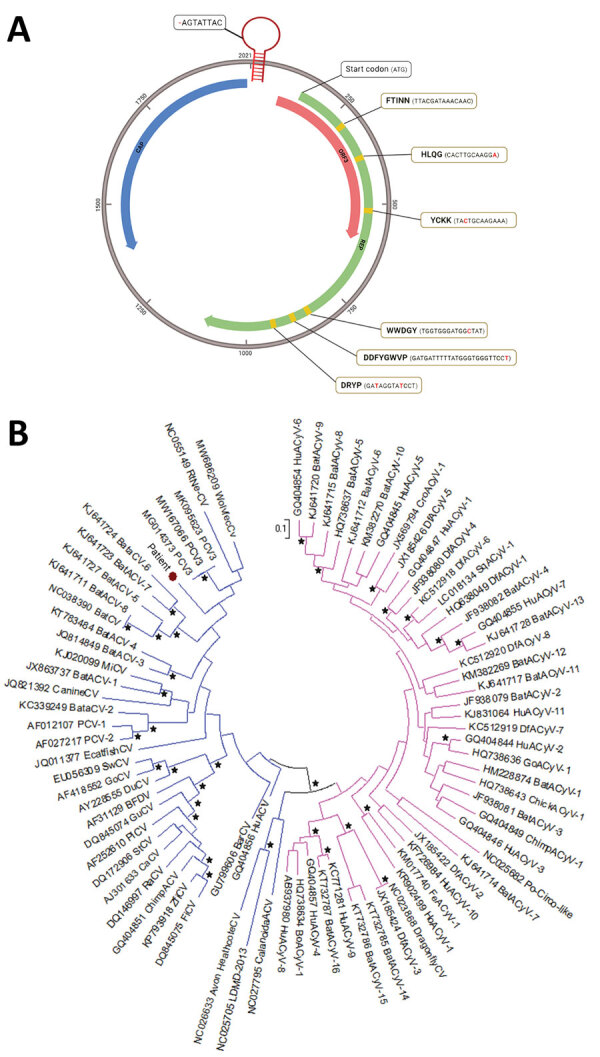
Genomic and phylogenetic analysis of putative novel virus, Circovirus parisii, from an immunocompromised patient with hepatitis, France, 2022. A) Full-length genome of C. parisii reconstructed from shotgun metagenomics (SMg) sequence analysis. The genome is a 2021-nt single-stranded circular DNA containing 3 predicted open reading frames (ORFs), including ORF1 (replicase, green), ORF2 (capsid protein, blue) and ORF3 (red). The stem-loop contains an AGTATTAC sequence (origin of replication) that misses 1 nt (red dash) compared with other circoviruses. An ATG start codon is located at the 5′ end of the replicase gene. The replicase gene contains 6 conserved motifs, represented with a yellow background (amino acid and nucleotide sequences), with silent substitutions in red. B) Phylogenetic analysis of the replicase gene of the Circoviridae family, including the newly discovered C. parisii (red dot), known circoviruses (blue), and known cycloviruses (pink). Bootstrap values >70% are indicated with black stars. Scale bar indicates substitutions per site.
