Figure.
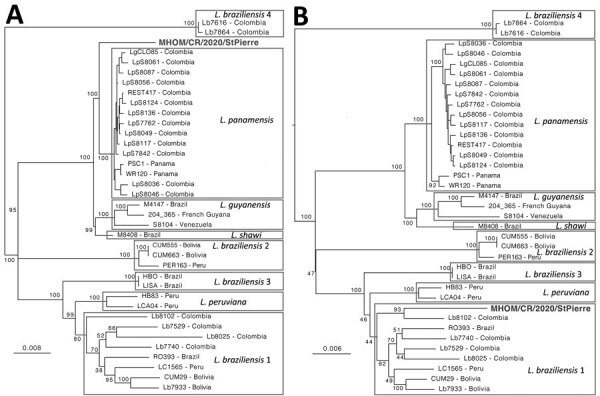
Midpoint rooted maximum-likelihood phylogenetic trees based on single-nucleotide polymorphisms called in chromosome 1 and the first 140kb of the telomeric region of chromosome 20 of a hybrid Leishmania parasite from Costa Rica. For each strain, sequences were composed based on concatenated single-nucleotide polymorphisms that were each coded by 2 base pairs, after which invariant sites were removed, resulting in 2,382 bp sequences for chromosome 1 and 3,015 bp sequences for chromosome 20. Consensus phylogenetic trees were generated from 1,000 bootstrap trees using IQ-TREE (http://www.iqtree.org) with 37 taxa (excluding L. naiffi and L. lainsoni strains) under the transversion with empirical base frequencies, ascertainment bias correction, and discrete gamma with 4 rate categories substitution model, which was the best-fit model revealed by ModelFinder as implemented in IQTREE. Branch support values are presented near each node following 1,000 bootstrap replicates; bootstrap values within the clade containing L. panamensis strains were omitted for clarity reasons. Scale bar indicates number of substitutions per site. Appendix 2) includes a description of the L. braziliensis 1–4 lineages.
