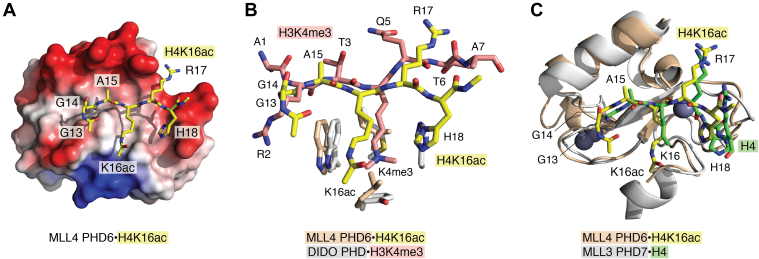
Figure 2.
Structural basis for the recognition of H4 by PHD fingers.A, the solution NMR structure of the MLL4 PHD6 finger in complex with H4K16ac peptide (PDB ID 6O7G). Electrostatic surface potential of the PHD6 finger is shown with blue and red colors representing surface positive and negative charges, respectively. The H4K16ac peptide is depicted as yellow sticks. B, overlay of the H4K16ac-binding site derived from the structure of the MLL4 PHD6 finger (wheat) in complex with H4K16ac peptide (yellow) (PDB ID 6O7G) and the H3K4me3-binding site derived from the structure of the DIDO PHD finger (gray) in complex with H3K4me3 peptide (dark pink) (PDB ID 4L7X). The K4me3-aromatic cage residues and the K16ac-hydrophobic groove residues are shown as sticks. Histone peptides residues are labeled. C, structural overlay of the MLL4 PHD6 finger (wheat) in complex with H4K16ac peptide (yellow) (PDB ID 6O7G) and the MLL3 PHD7 finger (gray) in complex with unmodified H4 peptide (green) (PDB ID 6MLC). MLL, mixed lineage leukemia, PHD, plant homeodomain.