Abstract
The twin-arginine translocation (Tat) pathway utilizes the proton-motive force to transport folded proteins across cytoplasmic membranes in bacteria and archaea, as well as across the thylakoid membrane in plants and the inner membrane in mitochondria. In most species, the minimal components required for Tat activity consist of three subunits, TatA, TatB, and TatC. Previous studies have shown that a polar amino acid is present at the N terminus of the TatA transmembrane helix (TMH) across many different species. In order to systematically assess the functional importance of this polar amino acid in the TatA TMH in Escherichia coli, we examined a complete set of 19-amino-acid substitutions. Unexpectedly, although the polar amino acid is preferred overall, our experiments suggest that it is not necessary for a functional TatA. Hydrophilicity and helix-stabilizing properties of this polar amino acid were found to be highly correlated with the Tat activity. Specifically, change in charge status of the amino acid side chain due to pH resulted in a shift in hydrophilicity, which was demonstrated to impact the Tat transport activity. Furthermore, we identified a four-residue motif at the N terminus of the TatA TMH by sequence alignment. Using a biochemical approach, we found that the N-terminal motif was functionally significant, with evidence indicating a potential role in the preference for utilizing different proton-motive force components. Taken together, these findings yield new insights into the functionality of TatA and its potential role in the Tat transport mechanism.
Keywords: twin-arginine translocation pathway, TatA, toroidal pore, alpha helix, hydrophobicity
The twin-arginine translocation (Tat) pathway, a protein transport machinery, is found in bacteria, archaebacteria, chloroplasts, and plant mitochondria. In bacteria, the Tat pathway is involved in many critical biological processes, including cell division, stress tolerance, and electron transport (1, 2, 3). In plants, the Tat pathway transports several proteins that are essential for photosynthesis across thylakoid membranes (4). In haloarchaea, which normally possess high cytoplasmic salt concentrations leading to faster protein folding, the Tat pathway serves almost 50% of secretome (5). Unlike the similarly ubiquitous Sec pathway, the Tat pathway is able to transport folded proteins (4) while requiring only the proton-motive force (pmf), with no contribution from NTP hydrolysis (6).
The Tat system in most species consists of three subunits, TatA, TatB, and TatC (1). It has been shown that all three subunits can form a complex, which works as a receptor for signal peptides of Tat substrates. After the receptor complex binds to the Tat substrate, more TatA subunits are recruited to the translocon. While TatB and TatC are present in a 1:1 stoichiometry in the complex, TatA is present at much higher stoichiometries (8- to 40-fold) (7, 8, 9), but see (10).
The structures of the three individual Tat subunits have been determined (11, 12, 13, 14). Both TatA and TatB possess a short transmembrane helix (TMH) followed by a hinge region, one or more amphipathic helices, and then a relatively unstructured C terminus. Conversely, TatC has six TMHs and exhibits a cupped hand shape. Remarkably, the Tat machinery transports folded proteins of different sizes without introducing significant ion leakage (15, 16). Recent work suggests that this occurs in the absence of a channel/pore-like structure, unlike the well-characterized Sec translocon (17). It has been proposed that the Tat pathway operates by forming transient lipid-lined toroidal pores in destabilized membranes, rather than utilizing a proteinaceous pore (15, 18, 19).
Previous studies have demonstrated that TatAs have a highly conserved short TMH (19), which could cause membrane destabilization and is critical to the overall Tat function (19, 20, 21). Such short TMHs make the TatA protein energetically unfavorable to span on the membrane bilayer. Furthermore, it has been shown that almost all TatA family proteins contain a polar residue at the N terminus of its TMH (22). In Escherichia coli, TatA has a glutamine (Q) at the eighth position (TMH: 6th–20th), while in pea, TatA has a glutamic acid (E) instead at the corresponding position. A cysteine-scanning study in E. coli TatA illustrated that cysteine substitution for the eighth glutamine (Q8C mutant) led to the complete loss of Tat activity, whereas cysteine substitutions for all the other residues in the TMH retained Tat transport activity similar to that of the wtTatA (23). Further studies showed that E. coli TatA asparagine substitution (Q8N) and Q8E mutants retained the ability to export trimethylamine N-oxide reductase (TorA) to the periplasm (23), while the alanine (Q8A) mutant was not able to transport either TorA (23) or CueO (24). However, another study showed that E. coli Q8A and Q8C mutants retained TorA transport capability when overexpressed (25). On the other hand, in pea or maize chloroplasts, it was shown that all of the 10th glutamic acid substitution variants, E10A, E10D, and E10Q, severely affected Tat transport in a biochemical complementation assay (26, 27). Interestingly, although the chloroplast TatA E10Q mutant exhibited extremely low Tat activity, the E. coli TatA Q8E mutant displayed high Tat transport activity, suggesting that residue preference was not identical for different species. Such different preference was also shown with chimeric pea/E. coli TatA derivatives (28). Furthermore, the Bacillus subtilis Tat system contains two sets of TatA/TatC subunits (TatAd/TatCd and TatAy/TatCy), and those TatAs actually lack this “conserved” polar residue in the TMH. Although extensive effort has been made to investigate the function of this “conserved” polar amino acid in TatA, the lack of a complete set of mutants and use of different transport assays has caused the role of this residue at the analogous position to remain an open question.
In this study, we analyzed the amino acid requirement for the eighth residue in E. coli TatA through a complete set of Q8 mutants. Both qualitative and quantitative assays were conducted to compare the activity between TatA mutants. Unexpectedly, we found that polar amino acids were preferred overall but actually not necessary for a functional TatA. However, the Tat activity was highly correlated with hydrophilicity and helix stability of the eighth residue in TatA. Furthermore, by sequence alignment and biochemical analysis, a four-residue motif at the N terminus of the TatA TMH was found to be functionally significant. These results expand our understanding of the potential function of TatA and provide new insight into the Tat transport mechanism.
Results
Polar amino acids at the eighth position in E. coli TatA TMH are preferred but not necessary for minimal Tat activity
To systematically assess the characteristic requirements of the eighth residue in E. coli TatA, Q8 was replaced by all the other 19 amino acids and their effects on Tat activity was measured. Membrane stability of TatA Q8 mutants was assessed by membrane isolation followed by carbonate wash, and all mutants displayed an integrated presence in the membrane (Fig. 1). Minimal Tat activity was subsequently screened by SDS growth assays (3) for each of the TatA Q8 mutants. All polar amino acid substitutions (Q8N, Q8S, Q8T, Q8D, Q8E, Q8H, Q8K, and Q8R) exhibited sufficient Tat activity to grow in LB medium containing 10% SDS (Fig. 2A). Surprisingly, TatA Q8A, Q8C, Q8G, and Q8P mutants, whose substituted amino acids do not possess strong polar side chains, also showed a growth activity in the same medium (Fig. 2, B and C). The rest of substitutions (Q8F, Q8I, Q8L, Q8M, Q8V, Q8W, and Q8Y) displayed no growth activity in LB medium containing 5% SDS, indicating a complete loss of Tat activity (Fig. 2C). An in vivo transport assay was implemented to further confirm the SDS growth assay result. In this assay, a FLAG-tagged E. coli native Tat substrate, SufI-FLAG, was overexpressed in the TatA Q8 mutants. After 2.5 h transport at 37 °C, a periplasmic fraction was isolated by treatment with EDTA/lysozyme and osmatic shock (29), a harsh method that minimizes the loss of periplasmic proteins. The precursor and mature forms of SufI-FLAG were separated by SDS-PAGE according to their size. This assay revealed that, despite their ability to grow in 10% SDS, the Q8A, Q8C, Q8D, Q8E, Q8G, Q8P, Q8R, and Q8T mutants displayed significantly lower Tat activity (<25%) compared with wildtype TatA (Fig. 2D).
Figure 1.
Membrane stability of the TatA Q8 substituted mutants. The membrane stability of the TatA Q8 mutants together with the wtTatA and ΔtatA were assessed by immunoblotting of the carbonate-treated membrane fraction from whole-cell extracts. Anti-TatA antibody was used to detect the presence of TatA in the membrane fractions.
Figure 2.
Polar residues at the eighth position of the E. coli TatA transmembrane helix are preferred but not necessary for minimal Tat activity.A–C, minimal Tat transport abilities of the TatA Q8 variants were evaluated by the growth performance in LB medium with indicated SDS concentrations. wtTatA served as the positive control, while ΔtatA acted as the negative control. Survival ratio denotes the ratio of the A600 of the cells grown in the presence of the corresponding SDS concentration to the A600 of the cells grown without SDS. Three biological replicates for each mutant were included in the experiments. D, Western blot results of the periplasmic fraction of in vivo transport assays with the TatA Q8 variants. Dilutions of the amount of transported SufI-FLAG in the wtTat cells were shown for the comparison of the transport performance in the TatA Q8 variants. p, precursor and m, mature. E, plot of helix stability (Δtm, the difference in denaturation temperature) versus side chain hydrophobicity (ΔtR, the difference in retention time in reversed-phase HPLC) of amino acids (32) colored according to their activity in the eighth position of TatA. Different levels of the Tat transport activities are denoted with the following rules: no Tat activity, no growth in the LB medium containing 5% SDS; low Tat activity, mutants who could grow in the LB medium with SDS but showed less than 50% transport activity than wtTat in the in vivo transport assay in (D); high Tat activity, mutants with more than 50% of the wtTat activity in (D). Amino acids with high Tat activity were clustered in the ranges of moderate helix stability and relatively high side chain hydrophilicity, which is boxed in pink.
Pulse-chase assays, representing a more fine-grained assessment of Tat transport activity (30), were conducted with all the functional TatA Q8 mutants to compare their transport rates. Figure 3A shows the quantitative analysis of the pulse-chase results (Fig. S1). An exponential plateau model, , was applied in data fitting. An estimated initial transport rate () derived from the fitting curve was used to compare the transport activity between the TatA Q8 mutants (Fig. 3B). Unexpectedly, the Q8H mutant exhibited the highest (228% of the [wtTatA]). Q8N, Q8S, and Q8E variants of TatA exhibited 58%, 37%, and 33% of the [wtTatA], respectively. However, Q8A, Q8C, Q8D, Q8G, Q8K, Q8P, and Q8R showed relatively lower transport rates (less than 25% of [wtTatA]). One unanticipated finding was that the Q8T, Q8D, and Q8R mutants, whose eighth glutamine was replaced by a polar amino acid, showed even lower transport rates than the poorly performing Q8A mutant. The results from pulse-chase assays were overall consistent with the in vivo transport assay except that Q8K performed relatively worse in the pulse-chase assay than in the in vivo transport assay.
Figure 3.
Quantitative analyses of the Tat activity in the TatA Q8 variants.A, plot of the transport kinetics of the TatA Q8 variants in pulse-chase experiments. Identities of wtTatA and TatA Q8 variants are shown in the right. Mature/total denotes the ratio of the mature band to the sum of the precursor and mature bands revealed by autoradiography of the samples from the pulse-chase experiment transporting SufI; chase time is denoted on the abscissa. Lines show best fits of an exponential model to the points arising from at least two biological replicates for each mutant. B, plot of the initial velocities (V0) in the TatA Q8 variants transporting SufI. Exact values of the V0 are shown above the bars. TatA Q8 variants are grouped based on classification of the amino acids.
In summary, polar amino acids are generally strongly favored at the eighth position in the E. coli TatA but are not absolutely necessary for a functional Tat translocon. Moreover, not all polar amino acid substitutions showed higher transport ability than nonpolar substitutions.
The Tat activity of Q8 mutants is correlated with the side chain hydrophilicity and helix-stabilizing properties of the substituted amino acid
Our data clearly show that polar amino acids are preferred overall in the TatA eighth position but are not strictly necessary to retain Tat activity (e.g. Q8A, Q8C, Q8G, Q8P). Moreover, some of the polar residue substitutions (i.e., Q8D, Q8R, and Q8T), even though they retained the minimal Tat activity required to grow in LB medium with SDS, displayed significantly lowered transport activity. Such results could not simply be explained by the binary categories, polar or nonpolar, of amino acids. In order to understand the functional importance of this position in the E. coli TatA, a further analysis of the amino acids’ preference is needed. On the one hand, previous studies showed that the overall hydrophobicity of the TatA TMH might affect TatA function in thylakoids (28). On the other, several recent studies demonstrated that the hydrophobic mismatch between the TatA TMH and membrane bilayer is critical to Tat function (19, 21, 31). To maintain such a hydrophobic mismatch, a relatively stable alpha helix structure is likely to be required. To assess these properties, the hydrophobicity and α-helix stability of each amino acid (32) were plotted together with the transport performance of the corresponding TatA Q8 mutants (Fig. 2E). Interestingly, a high correlation between Tat activity and the combination of hydrophilicity and α-helix stability was observed: (1) The Q8 mutants, which completely lost Tat activity, clustered at the top-right corner, suggesting that very high hydrophobicity and helix stability at this position are detrimental for Tat transport. (2) All mutants with high Tat activity were clustered in the middle ranges of hydrophilicity and helix stability (Fig. 2E, pink box). These observations point to the importance of a moderate hydrophilicity and α-helix stability of the eighth amino acid in E. coli TatA.
Charge status of the side chain of the polar amino acid affects the TatA function
Owing to the nature of polar amino acids with electrically charged side chains, previous studies have reported that the hydrophobicity of those amino acids changes dramatically under different pH conditions (33, 34). For instance, the side chain of histidine (pKa(His) = 6.04) is relatively hydrophobic (similar as Ala) at pH 7 (mostly uncharged), while it becomes one of the most hydrophilic side chains when the pH is lower than 5 (mostly charged). In this study, we hypothesized that a moderately hydrophilic side chain is required for the eighth residue in the E. coli TatA. To further investigate this hypothesis, a comparison of Tat transport rates between wtTatA and Q8H was made under different pH conditions. The pulse-chase experiments in Figure 3 showed that the Q8H mutant performed approximately 2-fold better than the wildtype TatA at pH 7.0 where most of (90%) the histidines in TatAs were presumably deprotonated. To assess whether a change in histidine hydrophobicity would alter activity of the Q8H mutant, we measured transport rates by pulse chase at pH 5.0, 6.0, and 8.0 (Fig. 4A). Figure 4 shows that wtTatA and Q8H had different behaviors when transporting SufI at different pH. First, the wtTatA and Q8H mutants exhibited optimal transport rates at different pH values (Fig. 4, B and C). Wildtype TatA displayed optimal transport activity at pH 5.0, while the optimal pH for the Q8H mutant was 7.0. However, their highest V0 values were comparable with each other (Fig. 4D). Second, [wtTatA] was significantly higher than [Q8H] at pH 5.0 (Fig. 4E), while [wtTatA] was significantly lower than [Q8H] at pH 7.0 (Fig. 4G). Third, wtTatA had a very similar as Q8H mutant at pH 8.0 (Fig. 4H) where 99% of the histidines were presumably deprotonated, a similar status as uncharged glutamine in wtTatA. In summary, the charged status of the histidine side chain, a model residue for comparison with the wtTatA, affects its transport.
Figure 4.
Comparison of the Tat activity of wtTat and Q8H mutant at different pH conditions.A, radioautography of the wtTatA and Q8H variants transporting SufI at different pHs. Samples from the pulse-chase experiment were subjected to SDS-PAGE. Polyacrylamide gels, 8 to 16%, were used to separate the mature SufI (m) from the precursor (p). Corresponding times of the chase steps are shown on top. Two replicates for each variant were included in this experiment. B and C, quantitative analyses of the transport performance of the wtTatA (B) and Q8H (C) variants based on the radioautography in (A). The same analysis approach was used as mentioned earlier. The pH condition in which the experiment was performed is indicated in the legend of the plot. D, plot of the initial velocities under different pH conditions between the wtTat and Q8H variants. Initial velocities of the wtTatA under different pH are denoted in black, and initial velocities of the Q8H variant under different pH are denoted in gray. E–H, comparison of the transport kinetics between the wtTatA and Q8H variants under the same pH (E, pH 5.0; F, pH 6.0; G, pH 7.0; H, pH 8.0), with the filled symbols representing the wtTatA and empty circles representing the Q8H. Other details are as described in the legend to Figure 3.
In aggregate, our experiments in the pH range 5.0 to 8.0 are consistent with the hypothesis that a functional Tat translocon requires that its TatA possesses a moderately hydrophilic amino acid at the eighth position. In addition, it demonstrates that not only hydrophilicity of different amino acids but also hydrophilicity of same amino acids under different charged status affects its ability to support Tat transport. Together these results provide important insights into understanding the function of the eighth residue in E. coli TatA (see Discussion).
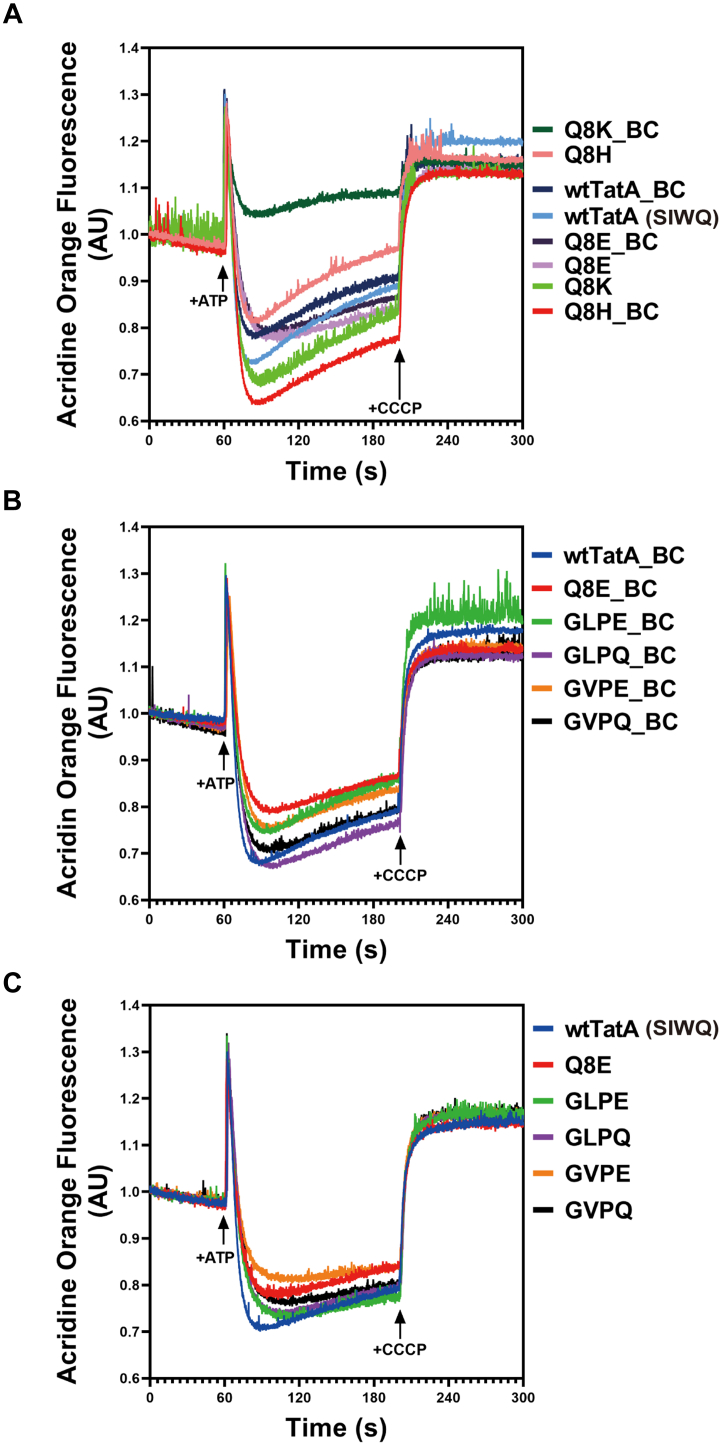
To further investigate how charged residues in TatA affect the overall Tat transport machinery, we determined how well the membrane can hold a pmf by measuring the ΔpH induced by ATP hydrolysis in inverted membrane vesicles (IMVs) membrane expressing corresponding mutated TatA proteins. According to their charge status at pH 7.0, Q8E, wtTatA, Q8H, and Q8K mutants were selected as examples of negatively, neutral, partially positively, and completely positively charged residues at the eighth position in E. coli TatA (Fig. 5A). Wildtype TatA and the Q8E mutant showed similar low membrane leakage outcomes in the presence or absence of the TatBC complex. However, Q8H and Q8K mutants exhibited significant different membrane leakage characteristics when the TatBC complex was present. Q8H, which showed the highest membrane leakage without TatBC, displayed less membrane leakage when TatBC was also expressed on the membrane. In contrast, the Q8K mutant showed the opposite behavior, exhibiting the highest ΔpH without TatBC but developing a dramatically high membrane leak when TatBC was present. That is to say, even though lysine and glutamic acid have similar hydrophilicity at pH 7.0 (Fig. 2E), their different charge status might affect how they interact with the TatBC complex, which results in different membrane leakage outcomes. The cause for this behavior is not clear and awaits further investigation.
Figure 5.
Membrane leakage profiles of TatA mutants. The ΔpH developed across the membrane was measured by the acridine orange quenching in the inverted membrane vesicles (IMVs). ATP, 4 mM, was added at 60 s to energize the IMV, and 10 μM carbonyl cyanide m-chlorophenyl hydrazone was added at 200 s to dissipate the proton gradient, resulting the recovery of the fluorescence. A, measurement of the ΔpH in the DADE (Tat knockout variant) IMVs with indicated overexpressed TatA Q8 variants alone or together with the constitutively expressed TatBC. B, measurement of the ΔpH in the DADE (Tat knockout variant) IMVs with indicated overexpressed TatA N-terminal motif variants alone. C, measurement of the ΔpH in the DADE (Tat knockout variant) IMVs with indicated overexpressed TatA N-terminal motif variants together with the constitutively expressed TatBC. AU, arbitrary unit.
Different species have different preferences for polar amino acids in the TatA TMH
Even though in E. coli TatA, the polar amino acid in the TMH is the uncharged glutamine, in plant Tat systems this position is usually occupied by a glutamic acid, a charged amino acid (35). Moreover, B. subtilis, a gram-positive bacterium, expresses two different kinds of TatA with glycine (TatAc) or serine (TatAy) at the corresponding position. To investigate if such amino acid choices are correlated to species, TatA sequences were aligned according to five categories: proteobacteria (N = 7534), actinobacteria and firmicutes (N = 6978), chloroplasts (N = 242), cyanobacteria (N = 149), and archaea (N = 151). A conserved 12-residue hydrophobic core was observed (9th–20th in Fig. 6, A–E) before the invariant “FG” motif at the end of TMH (19). In addition, there is a clear preference for particular amino acids in the TatA TMH at the eighth position with different species (Fig. 6F). (Here, we use the numbering from E. coli for simplicity. The exact number might be different in different species.) In proteobacteria (representing E. coli), histidine (50%) and glutamine (34%) are the top two choices. Interestingly, such choice preferences are consistent with our Tat activity results in that Q8H and wtTatA exhibited the highest Tat transport rates. In contrast, in the other two groups of bacteria, actinobacteria and firmicutes, even though histidine is still the top choice for TatA (46%), glutamine is present in only 2% of the species in these categories, and relatively more of them choose glutamic acid (32%) or glycine (10%) compared with proteobacteria. However, all of chloroplasts, cyanobacteria, and archaea TatAs have a highly conserved glutamic acid (>90%) at the eighth position. Remarkably, even though polar amino acids are present in TatA in most species, 1% of actinobacteria and firmicutes (N = 80) and 0.1% of proteobacteria (N = 13) possess an alanine at that position. This finding further confirms our experimental observations that polar residues at the eighth position are not indispensable for a functional TatA (Fig. S2, A and B). Such different preferences in different species suggest that a possible evolutionary pressure exists at this position for the Tat pathway to accommodate various external conditions.
Figure 6.
Different species have different preferences at the eighth position in TatA. Sequence logos of the TatA sequences alignment across (A) proteobacteria, (B) actinobacteria and firmicutes, (C) chloroplasts, (D) cyanobacteria, and (E) archaea. Numbers of sequences included in the corresponding categories were denoted as N. The N-terminal motifs were boxed as indicated. F, distributions of amino acids selected in the location equivalent to the eighth position of the E. coli TatA across different species. Percentage of species with the indicated residues were shown. Amino acids were colored as indicated to the right of the plot.
A four-residue functional motif at the N terminus of the TatA TMH is important for Tat activity
Besides the various preference of amino acids at the eighth position in different species, a conserved GxPE motif (the fifth to eighth position in Fig. 6, C, D and E) was also observed in chloroplasts, cyanobacteria, and archaea TatAs. In contrast, in proteobacteria, actinobacteria, and firmicutes, such “GxPE” motif was not obviously presented. Instead, a SIW(H/Q) motif was generally conserved in proteobacteria. To see if these motifs are correlated to different polar amino acids, TatA sequences were further examined based on the identity of the amino acid at the eighth position (Fig. S2). Fig. S2A clearly shows that proteobacteria favor “SIWH” and “SIWQ” (i.e., E. coli) at the N terminus of the TatA TMH. In addition, “GLPG,” “SITK,” and “S(I/L)WN” motifs were identified in proteobacteria as well. Interestingly, in E. coli TatE, “SITK” was at the corresponding location (the fifth to eighth residues). It has been reported that the TatE gene was found only in enterobacteria, a subset of proteobacteria (36). Accordingly, in this study, TatA or TatE sequences from enterobacteria were also aligned (Fig. S2F) to check if such motifs are also present in other TatE genes. Surprisingly, “SIWQ” and “SITK” were extremely conserved across TatA (N = 599) and TatE (N = 160), respectively, in enterobacteria. In addition, although “SIWH” was the most conserved motif in proteobacteria, gram-positive bacteria favored “xPWH” in the same position (Fig. S2B). These results suggest that the four-residue motif at the N terminus of the TatA TMH might play an important role in TatA function despite minor deviations in different species.
To assess whether the corresponding four-residue motifs are required for specific amino acids at the eighth position to exhibit higher Tat transport activity, we also substituted the fifth to eighth amino acids in E. coli TatA according to the above conserved motifs, including “GVPE” (a motif of chloroplasts TatAs), “GLPE” (a motif of cyanobacteria TatAs), and “SITK” (a motif of enterobacteria TatEs). Moreover, to comprehensively understand how the fifth to seventh amino acids affect TatA function, the fifth to eighth residues of E. coli TatA were also changed to “GVPQ” or “GLPQ” (Fig. 7).
Figure 7.
Comparison of the Tat transport activity with the TatA N-terminal motif mutants.A, membrane after carbonate washing and total extract samples were subjected to SDS-PAGE and immunoblotting with the α-TatA antibody. Molecular weight is shown on the left. B, periplasmic fractions (top) and total extract (bottom) of the cells in the in vivo transport assays were isolated. Precursor (p) and mature (m) proteins were labeled on the right, and molecular weight is shown on the left. C–E, plots of the transport kinetics for the N-terminal motif mutants in the pulse-chase experiment. At least two replicates were included for each mutant. F, Plot of the initial velocities (V0) of the N-terminal motif mutants in the pulse-chase experiments. V0 was obtained from the kinetic plots in (C–E). The same methodology was implemented as described before. Exact values of the initial velocities are displayed on top of the bar graph. Other details are as described in the legend to Figure 3.
A recent study has shown that a shorter TMH negatively affects TatA’s membrane stability (19). In terms of our new mutants, GVP(E/Q) and GLP(E/Q), it was possible that those mutants lost their ability to stably embed into the membrane since proline (immediately before the eighth residue) usually acts as a helix breaker (37) in protein secondary structure. Figure 7A shows that TatA in GVP(E/Q) and GLP(E/Q) mutants exhibited much lower membrane abundance compared with wtTatA or Q8E mutant, indicating that a proline residue severely affects the membrane insertion, even though it did not cause high membrane leakage in those mutants (Fig. 5, B and C). But to our surprise, the GVPE mutant displayed better performance than the Q8E mutant in the in vivo transport assay (Fig. 7B). Pulse-chase assays with those N-motif mutants were also conducted to quantitatively compare their transport rates. Both GLPQ and GVPQ displayed somewhat lower transport rates than wtTatA, suggesting that a disruption of the conserved “SIWQ” motif was deleterious to E. coli TatA function (Fig. 7C). However, GVPE and SITK mutants exhibited significantly higher transport rates than Q8E and Q8K mutants, respectively, which indicates that the evolutionally conserved motifs are functionally more suitable for certain amino acids at the eighth position in TatA (Fig. 7, D–F). Despite that replacing to conserved N-motifs was generally beneficial for Tat function, we also observed that the GLPE mutant led to significantly lower transport rates than both the GVPE and Q8E mutants. Such results demonstrate that changes in a single amino acid in the N-terminal motif (even from one hydrophobic residue to another hydrophobic residue) strongly affected the final TatA function. This confirms our hypothesis that the N terminus of TatA, and not just the well-known polar amino acid, determines the Tat transport activity. Furthermore, it implies that the combinations of these amino acids at the N terminus of the TatA TMH are potentially tuned to their own membrane environments, and those different motifs could be a strategy to adjust to diverse and subtle biological membrane properties.
The conserved GVPE motif from chloroplast TatA might be responsible for preference of utilization of ΔpH
Even though the GVPE mutant performed significantly better than the Q8E mutant (Fig. 7F), it was still less effective than the wild-type SIWQ motif in the E. coli TatA.
This result does not point to an explanation of why most chloroplasts and cyanobacteria possess a glutamic acid at the eighth position in their TatAs. It was previously shown that the chloroplast Tat system is highly dependent on the membrane ΔpH (38), while the E. coli Tat system does not strictly require a ΔpH for active transport (39). These energetic preferences are also consistent with their own respective tendencies to partition their pmfs between a ΔpH and a Δψ (40, 41). To investigate if the invariant motif in chloroplast TatA is related to this energetic preference, IMVs, the bacterial counterpart to chloroplast thylakoids, were made from wtTatA, Q8E, and GVPE mutants. A previous study (39) used IMVs to test Tat transport ability powered by different components of the pmf. In our hands also, the IMVs made with wildtype E. coli Tat displayed a higher transport ability in the presence of nigericin, which presumably dissipated the ΔpH and increased Δψ. Figure 8 shows that wtTatA and the Q8E mutant displayed consistently higher performance in the presence of nigericin. However, the GVPE mutant exhibited significantly lower transport capability when the ΔpH was dissipated. These results suggest that the GVPE mutant relied more on ΔpH for transport than wtTatA or the Q8E mutant.
Figure 8.
TatA N-terminal mutants display differences in energy utilization preference. Comparison of the inverted membrane vesicle transport of SufI among wtTatA, TatA Q8E (SIWE), and TatA GVPE mutants in response to 10 μM nigericin or 10 μM valinomycin treatment. The control groups received equivalent amounts of EtOH and were set to 100%. The exact percentage of transport in the experimental groups is shown above the bars. The error bar represents the standard error of the mean. Each treatment group per mutant was performed in four replicates; see Fig. S3. Data were analyzed by t tests. The significance level is denoted on the top of columns. ∗∗p ≤ 0.01; ∗p ≤ 0.05; ns, p > 0.05.
Discussion
An incomplete set of E. coli TatA Q8 mutants was a limitation to a full understanding of the function of the “conserved” polar amino acid in the TatA TMH. In this study, we systematically analyzed the Tat transport ability of all the Q8 mutants where the eighth glutamine of E. coli TatA was substituted by the other 19 amino acids. Three assays were conducted to compare the transport rates between different mutants, qualitatively and quantitatively. The transport ability results of our TatA Q8 mutants showed that a polar amino acid at the eighth position of TatA is preferred but not strictly necessary for a functional TatA (Figs. 2 and 3). Rather, a strong correlation between Tat transport activity and the hydrophilicity and helix stability of the eighth residue was observed (Fig. 2E), such that a residue possessing a moderate hydrophilicity and helix stability was preferred for TatA to retain high transport capability. Moreover, by altering the environmental pH in our transport assay, we demonstrated that the Q8H mutant, which contains a protonatable amino acid substitution, exhibited significantly lower transport ability under conditions in which this residue was charged, that is, where the hydrophilicity of the eighth residue was much higher than the hypothesized required range (Fig. 4). Furthermore, by analyzing alignments of native TatA sequences, we found that species from different categories have different preferred amino acids at the eighth position correlating with conserved four-residue motifs at the N terminus of TMH (Fig. 6). Such four-residue motifs were further tested in E. coli TatA and proved to have significant effect on the transport ability (Fig. 7). Interestingly, these motifs were correlated with predominant usage of the ΔpH or Δψ components of the pmf (Fig. 8). In the congregate, these results provide new perspectives through which to understand the functional significance of the eighth residue in TatAs.
In Figure 1, it can be seen that substitutions at Q8 of TatA did not strongly affect its membrane association, and those mutants showed various transport activities. In contrast, some substitutions from position five to eight of TatA significantly diminished membrane insertion, although they still retained decent transport activity. Thus, no strong correlation was observed between membrane association and transport activity.
Our finding that the hydrophilicity, but not polarity, of the eighth amino acid is critical for TatA supports the idea that this residue’s ability to respond to membrane potential or to electrically interact with other polar amino acids in other Tat subunits is not necessary for the fundamental transport mechanism. Given that E. coli TatA lacks a charged polar amino acid while chloroplast TatAs have a charged residue at the corresponding position, the possibility that bacteria and thylakoids might use different mechanisms in terms of TatA function has been discussed (42). It has been debated whether an active shift of protonation states of the glutamic acid in the chloroplasts TatA is required for a functional transporter (1, 42). However, this cannot be the case since many TatAs do not have this protonatable residue, and we find that even Ala in this position confers some activity. Instead, the requirement appears to be for a residue with a moderate hydrophilicity and helix stability. In that context, the pKa of the side chain of glutamic acid roughly matches the thylakoid’s lumen pH in the light (43), and this changes the overall hydrophilicity of the amino acid (33) to the moderate range. Such an explanation is further supported by two observations. First, it has been shown that thylakoid Tat transport operates with a delay between the light illumination and transport event (a sigmoidal transport curve), even at saturating TatA concentrations (44). Such a delay cannot be completely attributed to the translocon assembly step and may also be caused by the time required to develop a lumen pH low enough to modulate the Glu hydrophilicity. Second, our sequence alignments revealed that nine archaea species have TatAs with an aspartic acid (Fig. S2E) at the corresponding position whose side chain’s pKa is even lower than that of glutamic acid. Interestingly, we found by analyzing the living environments for those species that all of them live in extreme acidic environments (Table S1). A random sample of archaea species with E8 did not reveal any acidic extremophiles. While these observations do not explain why the TatAs have acidic amino acids that require neutralizing in the first place, they do provide examples of how the Tat system might be altered to accommodate to different physiological conditions. Moreover, a potential TatA–TatC interaction through a hydrogen bond between TatA Q8 and TatC Q215 was suggested from coevolution and modeling data (24). While this interaction is possibly important for optimized Tat activity in different species, it is unlikely to be essential for the Tat transport mechanism for three reasons. First, TatA Q8G and Q8A mutants, which cannot form such a hydrogen bond, still maintained reasonable transport activity in our experiments (Figs. 2 and 3). Second, there are multiple species from gram-positive bacteria that possess a non-hydrogen-bonding glycine at the corresponding position in their TatA proteins (Fig. S2). Finally, Alcock et al. showed that a TatC Q215A mutant still maintained significant Tat activity, even though no hydrogen bond between this TatC position and the TatA Q8 was possible. In addition, it is not the residue hydrophilicity only that changes when we substitute the eighth glutamine to other amino acids or when we change the pH environment. It is possible that other properties of a certain amino acid are also involved in the transport process, which were underestimated in our assay. However, due to the nature of amino acids, it is very difficult to solely investigate one property of the side chain while keeping others invariant.
Our experiments suggest that the creation of a relatively unstable N terminus structure by placing a polar eighth residue within the hydrophobic TatA TMH is likely significant. Such a residue would be expected to be located at or near the membrane/water interface, thereby biasing the position of the N terminus of the TMH close to the membrane surface (12). Meanwhile, two recent studies reported that the E. coli TatA TMH is tuned to be 15 residues long to keep the hydrophobic mismatch while also maintaining membrane integrity (19, 21) Also, it has been shown that TatAs have a highly conserved 12-residue hydrophobic core (9th–20th in E. coli TatA) in the TMH (19, 21). Together these observations indicate that the TatA TMH could be as short as 12 amino acids while the eighth residue is transiently exposed to the aqueous environment. This might allow membrane bilayer rupture (i.e., toroidal pore formation), with concomitant transport of a substrate held at the site. Our data herein demonstrated that, if the eighth residue was too hydrophobic (Fig. 2E), Tat transport was completely blocked, even though the hydrophobic mismatch between TatA TMH and the membrane bilayer was still maintained. This suggests that a 15-residue-long TMH is required but insufficient for active Tat transport and a transition from that length to an even shorter TMH might be functionally important.
We not only showed that the eighth residue is critical but also found that a changing four-residue motif at the N terminus of the TatA TMH affects the overall TatA function (Figs. 6 and 7). This motif was perhaps not identified in previous studies for two reasons. First, the functional N terminus motif was not obviously conserved across different species. For example, in chloroplasts and cyanobacteria, a GxPE motif was nearly invariant at the N terminus of the TatA TMH. However, this motif is not present in proteobacterial species. Second, the identified N-terminal motifs showed strong correlation with the eighth residue in the TatA TMH. For instance, in proteobacteria, “SIW” was conserved in the fifth to seventh positions when the eighth residue was glutamine or histidine, whereas this motif was not conserved when lysine or glycine was at the eighth position (Fig. S2). Even though we still do not understand the explicit roles of the conserved motifs, the amino acid choices therein likely provide clues about the function of the N terminus of the TatA TMH. For instance, a tryptophan was observed immediately before H8 or Q8 in proteobacteria and in two groups of gram-positive bacteria. One question would be what are the special features of tryptophan that cause it to be conserved in this context. Tryptophan is normally considered to be a nonpolar amino acid in biochemistry textbooks. However, it possesses a highly polarizable indole group in the side chain. This results in tryptophan being highly abundant at the water–lipid interface and acting as an anchor to stabilize membrane protein segments (45, 46). Moreover, the indole group could respond to an electric field and lower the conformational energy of the TMH were it to algin with the electrical force lines (47). Thus, the tryptophan might help TatA embed in the membrane and serve as an anchor near the interface. In addition, we noticed that a proline residue is highly conserved at position 7 when a glutamic acid is in position 8 in TatAs. Proline is generally considered a helix breaker in soluble proteins (48). However, it has been suggested that proline would not disrupt an alpha-helix in a membrane environment (49, 50). Moreover, it has been shown that the hinge-bending motions of proline residue are likely to play a significant role in catalysis and signal transduction (51, 52). Such a hinge-forming function at the N terminus of the TatA TMH is consistent with our hypothesis that a transition to an even shorter TMH might be beneficial for the Tat mechanism. In addition, according to the TatA NMR structure (11, 12), this four-residue motif presents in the first coil of the TMH. Ser6 and Gln8 orientate oppositely than Ile6 and Trp7, suggesting a weak local amphipathic topology. Moreover, other four-residue motifs in chloroplasts and gram-positive bacteria are also arrayed this way, even though they have different combinations of four residues at those positions. It is possible that this local amphipathic character assists in the reported motion of the TMH into and out of the membrane during transport (53), or, as suggested by (31), in the putative reorientation of the TMH to lie parallel to the plane of the membrane.
Incorporating our new findings related to the locally unstable and amphipathic properties of the N terminus of TatA, we propose a speculative new model for the mechanism of Tat pathway (Fig. 9). In this model, presented here to stimulate discussion: (i) Before substrate binding, TatA, TatB, and TatC form an oligomeric structure that serves as the receptor complex for the signal peptide of Tat substrates (24). At this stage, relatively few TatA proteins are associated with the TatBC complex. (ii) After substrate binding and (iii) in the presence of a pmf, the receptor complex undergoes a conformation change that attracts more TatA proteins (54). Unlike previous models (55), we take the view of (21) and propose these additional TatA proteins will be initially recruited into a noncircular structure, perhaps arrayed as the double line illustrated. (iv) As more TatA proteins are recruited to the translocon, the hydrophobic mismatch, unstable N terminus structure, and the membrane-thinning action of the pmf (56, 57, 58, 59) cause the double line to separate and join with subunits in adjacent lines to form a ring with a tight-fitting toroidal pore around the substrate (v). (vi) The concomitant movement of protons during substrate translocation (60) results in a local dissipation of the pmf, and this in turn returns the membrane to a thickness that favors the bilayer, causing dispersal of TatA from the translocation site and return of the translocon to its resting condition. While certain aspects of this model are admittedly speculative, we believe it captures many essential properties of Tat transport discerned from experiments. Future studies probing the biophysical properties of the membranes at the site of protein translocation are likely to shed further light on the mechanism of this enigmatic protein transport system.
Figure 9.
Schematic diagram for the proposed Tat transport mechanism. In this diagram, a view of the proposed Tat transport process is shown from above the membrane. Side view of the TatA interfaces is shown on the right of step 4. Blue/gray circles represent lipid head groups.
Experimental procedures
Strain and plasmid constructions
E. coli strain DADE-A (MC4100, ΔtatABCE, arabinose resistance) was used in this study (61). For TatA variants in pTat101 (62), which is a low-copy plasmid expressing the TatABC constitutively, the corresponding single and multiple amino acid substitutions were introduced by QuikChange site-directed mutagenesis (NEB, Phusion High-Fidelity PCR Kit) followed by KLD (Kinase, Ligase, and DpnI) digestion (NEB). Detailed primers used for TatA variant construction can be found in Table S2. For TatA variants in pBAD22, which is a vector with an arabinose-inducible araBAD operon, indicated TatA alleles, together with the wildtype TatBC, were amplified from the respective constructs in pTat101. For TatA single Q8 substitution, the forward primer TatA_gopBAD22_F (5′-ACC ACA GAG GAA CAT GTA TGG GTG GTA TCA GTA TTT G-3′) was used. For TatA multiple substitutions, the forward primer TatAnmotif_gopBAD22_F (5′- CTA CCA CAG AGG AAC ATG TAT GGG TGG TAT C-3′) was used. Reverse primer TatABC_gopBAD22_R (5′-TCT AGA GGC GGT TGA ATT TAT TCT TCA GTT TTT TCG C-3′) was used in both cases. Empty vectors were constructed using the primers pBAD22delABC_F (5′-ATT CAA CCG CCT CTA GAG-3′) and pBAD22delABC_R (5′-ACA TGT TCC TCT GTG GTA G-3′). Fragments and vector were assembled using the Gibson Assembly approach (NEBuilder HiFi DNA Assembly Master Mix). For TatA variants in pBAD33, which is a vector with an arabinose-inducible araBAD operon, a 6× His tag was first added at the C terminus of TatA by using the primers pBAD33His_F (5′- CATCATCATCACCACCACTAATGGC-3′) and pBAD33His_R (5′-TGATGATGACCCACCTGCTCTTTATCGTG-3′). Subsequently, TatA mutations were introduced by QuikChange site-directed mutagenesis as described above. pQE80l (SufI-FLAG) for the in vivo transport assay was constructed as described (63). pNR14 and pNR42, two plasmids used in pulse-chase experiments, were as described (64, 65). All plasmids were confirmed by Sanger sequencing. More detailed information on plasmid constructions can be found in Table S3.
Sequence alignments and statistical analyses
A total of 15,054 TatA sequences were downloaded from the identical protein groups in GenBank (NCBI). Sequences were then divided into five subgroups based on taxonomy (7534 from proteobacteria, 6978 from firmicutes and actinobacteria, 149 from cyanobacteria, 242 from chloroplast, and 151 from archaea) and were subsequently subjected to multiple sequence alignments within each group using MAFFT (66). Sequence logos corresponding to each group were generated using RStudio (Ver. 1.4.1103) with ggseqlogo package (67). For the interest of this project, sequences were further grouped by the identity of the residue aligned to the eighth position in E. coli TatA, and the corresponding sequence logos were generated. A stacked bar chart summarizing the distribution of the identity of residues in this position in each taxonomical group was plotted using GraphPad Prism version 8.2.1 for Windows (GraphPad Software).
Liquid SDS growth assay
Overnight bacterial cultures subcultured with an absorbance (A600) of 0.002 were grown in the Luria–Bertani (LB) medium containing 0%, 5%, or 10% SDS, respectively. After cells were incubated at 37 °C with shaking for 5 h, the final A600 was measured. For each mutant, the survival ratio was calculated by its final A600 at the indicated SDS concentration to the final A600 in the LB medium without SDS.
In vivo transport assay and cell fractionation
Corresponding mutant plasmids derived from pTat101 were cotransformed with pQE80l (SufI-FLAG), which is under the control of the T5 promoter, into the DADE-A strain. Overnight cultures were diluted in a fresh LB medium at a ratio of 1:100 and grown at 37 °C until A600 reached 0.5. A final concentration of 1 mM IPTG (isopropyl β-D-1-thiogalactopyranoside) was added to induce the expression of SufI-FLAG. After culturing at 37 °C for another 2.5 h, 3 ml of the cells with an A600 equivalent to 1 were fractionated using the EDTA/lysozyme/cold osmotic shock method described before (29). Periplasmic fractions were collected by adding an equal volume of 2× SDS sample buffer, which were then subjected to SDS-PAGE. Membrane fractions were washed in 10 mM Na2CO3 for 1 h at 4 °C with gentle shaking. Subsequently, samples were ultracentrifuged at 120,000g for 45 min at 4 °C. Pellets were resuspended with 2× SDS sample buffer and collected as the membrane fractions.
SDS-PAGE and Western blot
For periplasmic fractions, samples were immunoblotted with α-FLAG (GenScript) followed by HRP-conjugated α-mouse antibodies (Santa Cruz Biotechnology). For membrane fractions, after transfer of the proteins to the PVDF membranes, PVDF membranes were cut and immunoblotted with α-TatA antibody followed by HRP-conjugated α-rabbit antibody (GenScript). Proteins were visualized using ProSignal Pico ECL Western Blotting detection kit (Genesee Scientific).
Pulse-chase experiment and autoradiography
The pulse-chase experiment protocol was modified from (65). Tat variants carrying pNR14 and pNR42 were grown overnight in LB media at 30 °C, which were then subcultured in fresh LB media the next day for 1.5 h at 30 °C. Subsequently, cells were harvested and normalized to the equivalent of 0.5 ml cells with A600 = 0.2. Cells were washed with 0.5 ml 1× M9 medium (diluted from 10× M9 medium supplemented with 0.1 mM CaCl2, 0.002% thiamine, 2 mM MgSO4, and 0.01% of the 18 amino acids excluding methionine and cysteine, pH 7.0). Cells were resuspended in 2.5 ml M9 medium and cultured for another hour at 30 °C. Then, cells were transferred to 42 °C for 15 min to induce T7 polymerase from pNR42. Rifampicin, 400 μg/ml, was added to inhibit the E. coli endogenous RNA polymerase, and cells were grown at 42 °C for another 10 min. Subsequently, cells were grown at 30 °C for another 20 min before being transferred to the indicated temperature until the completion of the experiment. If needed, pH was adjusted by 1 M HCl or 1 M KOH immediately after this step. Bacterial cultures were then incubated at the indicated temperature for 5 min. [35S] methionine, 0.025 mCi (PerkinElmer Inc, NEG772002MC), was added for the pulse step, and 750 μg/ml of unlabeled cold methionine was added after 5 min. A volume of 300 μl of the culture was taken at the indicated time point, and transport was quenched by freezing in liquid nitrogen. Samples were subsequently thawed on ice, centrifuged, and resuspended with 2× SDS sample buffer followed by SDS-PAGE and autoradiography. Results were quantified using ImageJ software.
Statistical analysis of the pulse-chase experimental results
Mature-to-total (sum of precursor and mature) data from the pulse-chase experiment were plotted against each time point, which were then fitted with the exponential plateau model deriving from the first-order reaction model, , using GraphPad Prism. was defined as the maximum of the mature-to-total value, and represents the initial mature-to-total value at the end of the pulse. t represents time in minutes. The initial velocity (), representing the transport rate when , was obtained by , with the units of the reciprocal minutes.
IMVs preparation
Tat proteins expression and IMVs preparation were performed as described (39), with the following modifications. Cells were resuspended with the same lysis buffer as described, which was then passed through a French press at ∼ 6000 psi once. Subsequently, cells were centrifuged at 30,000 × g for 20 min at 4 °C. Supernatants were then centrifuged at 140,000 × g for 1 h. Pellets were resuspended with IMV storage buffer (10 mM Tris-HCl, pH 7.5, 1 mM MgSO4, 1 mM KCl, 40% glycerol). IMVs were stored at −80 °C.
Proton leakage measurement
IMVs were made from the DADE-A cells expressing the 6× His-tagged wildtype or TatA variants in pBAD33 alone or together with the pTat101(ΔtatA). Acridine orange fluorescence-quenching assays were carried out using the Fluorolog-3 spectrofluorometer (HORIBA Scientific, model No. FL3-22). IMVs were added to the final concentration equivalent to A280 = 0.375 to the reaction mix containing 1× TE buffer (25 mM MOPS, 25 mM MES, 5 mM MgCl2, 50 mM KCl, 200 mM sucrose, and 57 μg/ml bovine serum albumin, pH 7.0), ATP regeneration system (2.9 mM phosphocreatine, 0.29 mg/ml creatine kinase), and 2 μM acridine orange. The reaction mix was incubated for 5 min with gentle stirring at 37 °C before measurement. The fluorescence of acridine orange was recorded at λex = 494 nm (slit = 1 nm) and λem = 540 nm (slit = 5 nm) per 0.1 s. ATP, 4 mM, was added at 60 s, and 10 μM of carbonyl cyanide m-chlorophenyl hydrazone was added at 200 s to dissipate the proton gradient.
Data availability
All data are contained within the article.
Supporting information
This article contains supporting information (68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78).
Conflict of interests
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
B. H., W. Z., S. M. T. conceptualization; B. H., W. Z., S. M. T. methodology; W. Z. software; B. H. validation; B. H., W. Z. formal analysis; B. H., W. Z., S. M. T. investigation; B. H., W. Z. writing – original draft; B. H., W. Z., S. M. T. writing – review & editing; B. H., W.Z . visualization; S. M. T. supervision; S. M. T. funding acquisition.
Funding and additional information
We gratefully acknowledge support from the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy through Grant DE-SC0020304 to S. M. T.
Reviewed by members of the JBC Editorial Board. Edited by Karen Fleming
Supporting information
References
- 1.Berks B.C. The twin-arginine protein translocation pathway. Annu. Rev. Biochem. 2015;84:843–864. doi: 10.1146/annurev-biochem-060614-034251. [DOI] [PubMed] [Google Scholar]
- 2.Gohlke U., Pullan L., McDevitt C.A., Porcelli I., Leeuw E de, Palmer T., et al. The TatA component of the twin-arginine protein transport system forms channel complexes of variable diameter. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10482–10486. doi: 10.1073/pnas.0503558102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ize B., Stanley N.R., Buchanan G., Palmer T. Role of the Escherichia coli Tat pathway in outer membrane integrity. Mol. Microbiol. 2003;48:1183–1193. doi: 10.1046/j.1365-2958.2003.03504.x. [DOI] [PubMed] [Google Scholar]
- 4.Clark S.A., Theg S.M. A folded protein can be transported across the chloroplast envelope and thylakoid membranes. Mol. Biol. Cell. 1997;8:923–934. doi: 10.1091/mbc.8.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghosh D., Boral D., Vankudoth K.R., Ramasamy S. Analysis of haloarchaeal twin-arginine translocase pathway reveals the diversity of the machineries. Heliyon. 2019;5 doi: 10.1016/j.heliyon.2019.e01587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun N.A., Davis A.W., Theg S.M. The chloroplast Tat pathway utilizes the transmembrane electric potential as an energy source. Biophys. J. 2007;93:1993–1998. doi: 10.1529/biophysj.106.098731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leake M.C., Greene N.P., Godun R.M., Granjon T., Buchanan G., Chen S., et al. Variable stoichiometry of the TatA component of the twin-arginine protein transport system observed by in vivo single-molecule imaging. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15376–15381. doi: 10.1073/pnas.0806338105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mori H., Summer E.J., Cline K. Chloroplast TatC plays a direct role in thylakoid ΔpH-dependent protein transport. FEBS Lett. 2001;501:65–68. doi: 10.1016/s0014-5793(01)02626-6. [DOI] [PubMed] [Google Scholar]
- 9.Sargent F., Gohlke U., Leeuw E de, Stanley N.R., Palmer T., Saibil H.R., et al. Purified components of the Escherichia coli Tat protein transport system form a double-layered ring structure. Eur. J. Biochem. 2001;268:3361–3367. doi: 10.1046/j.1432-1327.2001.02263.x. [DOI] [PubMed] [Google Scholar]
- 10.Hauer R.S., Schlesier R., Heilmann K., Dittmar J., Jakob M., Klösgen R.B. Enough is enough: TatA demand during tat-dependent protein transport. Biochim. Biophys. Acta. 2013;1833:957–965. doi: 10.1016/j.bbamcr.2013.01.030. [DOI] [PubMed] [Google Scholar]
- 11.Hu Y., Zhao E., Li H., Xia B., Jin C. Solution NMR structure of the TatA component of the twin-arginine protein transport system from gram-positive bacterium Bacillus subtilis. J. Am. Chem. Soc. 2010;132:15942–15944. doi: 10.1021/ja1053785. [DOI] [PubMed] [Google Scholar]
- 12.Pettersson P., Ye W., Jakob M., Tannert F., Klösgen R.B., Mäler L. Structure and dynamics of plant TatA in micelles and lipid bilayers studied by solution NMR. FEBS J. 2018;285:1886–1906. doi: 10.1111/febs.14452. [DOI] [PubMed] [Google Scholar]
- 13.Rollauer S.E., Tarry M.J., Graham J.E., Jääskeläinen M., Jäger F., Johnson S., et al. Structure of the TatC core of the twin-arginine protein transport system. Nature. 2012;492:210–214. doi: 10.1038/nature11683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y., Wang L., Hu Y., Jin C. Solution structure of the TatB component of the twin-arginine translocation system. Biochim. Biophys. Acta BBA - Biomembr. 2014;1838:1881–1888. doi: 10.1016/j.bbamem.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 15.Asher A.H., Theg S.M. Electrochromic shift supports the membrane destabilization model of Tat-mediated transport and shows ion leakage during Sec transport. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2018122118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teter S.A., Theg S.M. Energy-transducing thylakoid membranes remain highly impermeable to ions during protein translocation. Proc. Natl. Acad. Sci. U. S. A. 1998;95:1590–1594. doi: 10.1073/pnas.95.4.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsirigotaki A., De Geyter J., Šoštarić N., Economou A., Karamanou S. Protein export through the bacterial Sec pathway. Nat. Rev. Microbiol. 2017;15:21–36. doi: 10.1038/nrmicro.2016.161. [DOI] [PubMed] [Google Scholar]
- 18.Brüser T., Sanders C. An alternative model of the twin arginine translocation system. Microbiol. Res. 2003;158:7–17. doi: 10.1078/0944-5013-00176. [DOI] [PubMed] [Google Scholar]
- 19.Hao B., Zhou W., Theg S.M. Hydrophobic mismatch is a key factor in protein transport across lipid bilayer membranes via the Tat pathway. J. Biol. Chem. 2022 doi: 10.1016/j.jbc.2022.101991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou B., Heidrich E.S., Mehner-Breitfeld D., Brüser T. The TatA component of the twin-arginine translocation system locally weakens the cytoplasmic membrane of Escherichia coli upon protein substrate binding. J. Biol. Chem. 2018;293:7592–7605. doi: 10.1074/jbc.RA118.002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mehner-Breitfeld D., Ringel M.T., Tichy D.A., Endter L.J., Stroh K.S., Luensdorf H., et al. TatA and TatB generate a hydrophobic mismatch that is important for function and assembly of the Tat translocon in Escherichia coli. bioRxiv. 2021 doi: 10.1101/2021.05.26.445790. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hicks M.G., Leeuw E de, Porcelli I., Buchanan G., Berks B.C., Palmer T. The Escherichia coli twin-arginine translocase: conserved residues of TatA and TatB family components involved in protein transport. FEBS Lett. 2003;539:61–67. doi: 10.1016/s0014-5793(03)00198-4. [DOI] [PubMed] [Google Scholar]
- 23.Greene N.P., Porcelli I., Buchanan G., Hicks M.G., Schermann S.M., Palmer T., et al. Cysteine scanning mutagenesis and disulfide mapping studies of the TatA component of the bacterial twin arginine translocase. J. Biol. Chem. 2007;282:23937–23945. doi: 10.1074/jbc.M702972200. [DOI] [PubMed] [Google Scholar]
- 24.Alcock F., Stansfeld P.J., Basit H., Habersetzer J., Baker M.A., Palmer T., et al. Assembling the Tat protein translocase. eLife. 2016;5 doi: 10.7554/eLife.20718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warren G., Oates J., Robinson C., Dixon A.M. Contributions of the transmembrane domain and a key acidic motif to assembly and function of the TatA complex. J. Mol. Biol. 2009;388:122–132. doi: 10.1016/j.jmb.2009.02.060. [DOI] [PubMed] [Google Scholar]
- 26.Dabney-Smith C., Mori H., Cline K. Requirement of a tha4-conserved transmembrane glutamate in thylakoid Tat translocase assembly revealed by biochemical complementation. J. Biol. Chem. 2003;278:43027–43033. doi: 10.1074/jbc.M307923200. [DOI] [PubMed] [Google Scholar]
- 27.Fincher V., Dabney-Smith C., Cline K. Functional assembly of thylakoid ΔpH-dependent/Tat protein transport pathway components in vitro. Eur. J. Biochem. 2003;270:4930–4941. doi: 10.1046/j.1432-1033.2003.03894.x. [DOI] [PubMed] [Google Scholar]
- 28.Hauer R.S., Freudl R., Dittmar J., Jakob M., Klösgen R.B. How to achieve Tat transport with alien TatA. Sci. Rep. 2017;7:1–13. doi: 10.1038/s41598-017-08818-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petiti M., Houot L., Duché D. In: Bacterial Protein Secretion Systems: Methods and Protocols, Methods in Molecular Biology. Journet L., Cascales E., editors. Springer New York; New York, NY: 2017. Cell fractionation; pp. 59–64. [Google Scholar]
- 30.Palmer T., Berks B.C., Sargent F. In: Protein Secretion: Methods and Protocols, Methods in Molecular Biology. Economou A., editor. Humana Press; Totowa, NJ: 2010. Analysis of Tat targeting function and twin-arginine signal peptide activity in Escherichia coli; pp. 191–216. [DOI] [PubMed] [Google Scholar]
- 31.Stockwald E.R., Steger L.M.E., Vollmer S., Gottselig C., Grage S.L., Bürck J., et al. Length matters: Functional flip of the short TatA transmembrane helix. Biophys. J. 2022 doi: 10.1016/j.bpj.2022.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monera O.D., Sereda T.J., Zhou N.E., Kay C.M., Hodges R.S. Relationship of sidechain hydrophobicity and α-helical propensity on the stability of the single-stranded amphipathic α-helix. J. Pept. Sci. 1995;1:319–329. doi: 10.1002/psc.310010507. [DOI] [PubMed] [Google Scholar]
- 33.Kovacs J.M., Mant C.T., Hodges R.S. Determination of intrinsic hydrophilicity/hydrophobicity of amino acid side chains in peptides in the absence of nearest-neighbor or conformational effects. Pept. Sci. 2006;84:283–297. doi: 10.1002/bip.20417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mant C.T., Hodges R.S. Reversed-phase liquid chromatography as a tool in the determination of the hydrophilicity/hydrophobicity of amino acid side-chains at a ligand–receptor interface in the presence of different aqueous environments: i. Effect of varying receptor hydrophobicity. J. Chromatogr. A. 2002;972:45–60. doi: 10.1016/s0021-9673(02)00354-0. [DOI] [PubMed] [Google Scholar]
- 35.Mori H., Cline K. Post-translational protein translocation into thylakoids by the Sec and ΔpH-dependent pathways. Biochim. Biophys. Acta. 2001;1541:80–90. doi: 10.1016/s0167-4889(01)00150-1. [DOI] [PubMed] [Google Scholar]
- 36.Yen M.-R., Tseng Y.-H., Nguyen E.H., Wu L.-F., Saier M.H. Sequence and phylogenetic analyses of the twin-arginine targeting (Tat) protein export system. Arch. Microbiol. 2002;177:441–450. doi: 10.1007/s00203-002-0408-4. [DOI] [PubMed] [Google Scholar]
- 37.Chou P.Y., Fasman G.D. Empirical predictions of protein conformation. Annu. Rev. Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- 38.Cline K., Ettinger W.F., Theg S.M. Protein-specific energy requirements for protein transport across or into thylakoid membranes. Two lumenal proteins are transported in the absence of ATP. J. Biol. Chem. 1992;267:2688–2696. [PubMed] [Google Scholar]
- 39.Bageshwar U.K., Musser S.M. Two electrical potential–dependent steps are required for transport by the Escherichia coli Tat machinery. J. Cell Biol. 2007;179:87–99. doi: 10.1083/jcb.200702082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cruz J.A., Sacksteder C.A., Kanazawa A., Kramer D.M. Contribution of electric field (Δψ) to steady-state transthylakoid proton motive force (pmf) in vitro and in vivo. Control of pmf parsing into Δψ and ΔpH by ionic strength. Biochemistry. 2001;40:1226–1237. doi: 10.1021/bi0018741. [DOI] [PubMed] [Google Scholar]
- 41.Zilberstein D., Agmon V., Schuldiner S., Padan E. Escherichia coli intracellular pH, membrane potential, and cell growth. J. Bacteriol. 1984;158:246–252. doi: 10.1128/jb.158.1.246-252.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.New C.P., Ma Q., Dabney-Smith C. Routing of thylakoid lumen proteins by the chloroplast twin arginine transport pathway. Photosynth Res. 2018;138:289–301. doi: 10.1007/s11120-018-0567-z. [DOI] [PubMed] [Google Scholar]
- 43.Kramer D.M., Sacksteder C.A., Cruz J.A. How acidic is the lumen? Photosynth. Res. 1999;60:151–163. [Google Scholar]
- 44.Celedon J.M., Cline K. Stoichiometry for binding and transport by the twin arginine translocation system. J. Cell Biol. 2012;197:523–534. doi: 10.1083/jcb.201201096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Jesus A.J., Allen T.W. The role of tryptophan side chains in membrane protein anchoring and hydrophobic mismatch. Biochim. Biophys. Acta. 2013;1828:864–876. doi: 10.1016/j.bbamem.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 46.Sun H., Greathouse D.V., Andersen O.S., Koeppe R.E. The preference of tryptophan for membrane interfaces. J. Biol. Chem. 2008;283:22233–22243. doi: 10.1074/jbc.M802074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X.C., Li H. Interplay between the electrostatic membrane potential and conformational changes in membrane proteins. Protein Sci. 2019;28:502–512. doi: 10.1002/pro.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barlow D.J., Thornton J.M. Helix geometry in proteins. J. Mol. Biol. 1988;201:601–619. doi: 10.1016/0022-2836(88)90641-9. [DOI] [PubMed] [Google Scholar]
- 49.Li S.C., Goto N.K., Williams K.A., Deber C.M. Alpha-helical, but not beta-sheet, propensity of proline is determined by peptide environment. Proc. Natl. Acad. Sci. U. S. A. 1996;93:6676–6681. doi: 10.1073/pnas.93.13.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Senes A., Engel D.E., DeGrado W.F. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr. Opin. Struct. Biol. 2004;14:465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 51.S.P. Sansom M., Weinstein H. Hinges, swivels and switches: the role of prolines in signalling via transmembrane α-helices. Trends Pharmacol. Sci. 2000;21:445–451. doi: 10.1016/s0165-6147(00)01553-4. [DOI] [PubMed] [Google Scholar]
- 52.Tieleman D.P., Shrivastava I.H., Ulmschneider M.R., Sansom M.S.P. Proline-induced hinges in transmembrane helices: possible roles in ion channel gating. Proteins Struct. Funct. Bioinform. 2001;44:63–72. doi: 10.1002/prot.1073. [DOI] [PubMed] [Google Scholar]
- 53.Aldridge C., Storm A., Cline K., Dabney-Smith C. The chloroplast twin arginine transport (Tat) component, Tha4, undergoes conformational changes leading to Tat protein transport. J. Biol. Chem. 2012;287:34752–34763. doi: 10.1074/jbc.M112.385666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aldridge C., Ma X., Gerard F., Cline K. Substrate-gated docking of pore subunit Tha4 in the TatC cavity initiates Tat translocase assembly. J. Cell Biol. 2014;205:51–65. doi: 10.1083/jcb.201311057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodriguez F., Rouse S.L., Tait C.E., Harmer J., Riso A.D., Timmel C.R., et al. Structural model for the protein-translocating element of the twin-arginine transport system. Proc. Natl. Acad. Sci. U. S. A. 2013;110 doi: 10.1073/pnas.1219486110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson M.P., Brain A.P., Ruban A.V. Changes in thylakoid membrane thickness associated with the reorganization of photosystem II light harvesting complexes during photoprotective energy dissipation. Plant Signal. Behav. 2011;6:1386–1390. doi: 10.4161/psb.6.9.16503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson M.P., Goral T.K., Duffy C.D.P., Brain A.P.R., Mullineaux C.W., Ruban A.V. Photoprotective energy dissipation involves the reorganization of photosystem II light-harvesting complexes in the grana membranes of spinach chloroplasts. Plant Cell. 2011;23:1468–1479. doi: 10.1105/tpc.110.081646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kirchhoff H., Hall C., Wood M., Herbstová M., Tsabari O., Nevo R., et al. Dynamic control of protein diffusion within the granal thylakoid lumen. Proc. Natl. Acad. Sci. U. S. A. 2011;108:20248–20253. doi: 10.1073/pnas.1104141109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murakami S., Packer L. Light-induced changes in the conformation and Configuration of the thylakoid membrane of Ulva and porphyra chloroplasts in Vivo. Plant Physiol. 1970;45:289–299. doi: 10.1104/pp.45.3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alder N.N., Theg S.M. Energetics of protein transport across biological membranes: a study of the thylakoid ΔpH-dependent/cpTat pathway. Cell. 2003;112:231–242. doi: 10.1016/s0092-8674(03)00032-1. [DOI] [PubMed] [Google Scholar]
- 61.Wexler M., Sargent F., Jack R.L., Stanley N.R., Bogsch E.G., Robinson C., et al. TatD is a cytoplasmic protein with DNase activity. No requirement for TatD family proteins in sec-independent protein export. J. Biol. Chem. 2000;275:16717–16722. doi: 10.1074/jbc.M000800200. [DOI] [PubMed] [Google Scholar]
- 62.Kneuper H., Maldonado B., Jäger F., Krehenbrink M., Buchanan G., Keller R., et al. Molecular dissection of TatC defines critical regions essential for protein transport and a TatB–TatC contact site. Mol. Microbiol. 2012;85:945–961. doi: 10.1111/j.1365-2958.2012.08151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang Q., Palmer T. Signal peptide hydrophobicity modulates interaction with the twin-arginine translocase. mBio. 2017;8:e00909–e00917. doi: 10.1128/mBio.00909-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sargent F., Stanley N.R., Berks B.C., Palmer T. Sec-independent protein translocation in Escherichia coli. J. Biol. Chem. 1999;274:36073–36082. doi: 10.1074/jbc.274.51.36073. [DOI] [PubMed] [Google Scholar]
- 65.Stanley N.R., Palmer T., Berks B.C. The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J. Biol. Chem. 2000;275:11591–11596. doi: 10.1074/jbc.275.16.11591. [DOI] [PubMed] [Google Scholar]
- 66.Rozewicki J., Li S., Amada K.M., Standley D.M., Katoh K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucl. Acids Res. 2019;47 doi: 10.1093/nar/gkz342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wagih O. ggseqlogo: a versatile R package for drawing sequence logos. Bioinformatics. 2017;33:3645–3647. doi: 10.1093/bioinformatics/btx469. [DOI] [PubMed] [Google Scholar]
- 68.Jones R.M., Johnson D.B. Acidithrix ferrooxidans gen. nov., sp. nov.; a filamentous and obligately heterotrophic, acidophilic member of the actinobacteria that catalyzes dissimilatory oxido-reduction of iron. Res. Microbiol. 2015;166:111–120. doi: 10.1016/j.resmic.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 69.Baumler D.J., Jeong K.-C., Fox B.G., Banfield J.F., Kaspar C.W. Sulfate requirement for heterotrophic growth of “Ferroplasma acidarmanus” strain fer1. Res. Microbiol. 2005;156:492–498. doi: 10.1016/j.resmic.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 70.Fütterer O., Angelov A., Liesegang H., Gottschalk G., Schleper C., Schepers B., et al. Genome sequence of Picrophilus torridus and its implications for life around pH 0. Proc. Natl. Acad. Sci. 2004;101:9091–9096. doi: 10.1073/pnas.0401356101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu L.-J., You X.-Y., Guo X., Liu S.-J., Jiang C.-Y. Metallosphaera cuprina sp. nov., an acidothermophilic, metal-mobilizing archaeon. Int. J. Syst. Evol. Microbiol. 2011;61:2395–2400. doi: 10.1099/ijs.0.026591-0. [DOI] [PubMed] [Google Scholar]
- 72.Golyshina O.V., Pivovarova T.A., Karavaiko G.I., Kondratéva T.F., Moore E.R., Abraham W.R., et al. Ferroplasma acidiphilum gen. nov., sp. nov., an acidophilic, autotrophic, ferrous-iron-oxidizing, cell-wall-lacking, mesophilic member of the Ferroplasmaceae fam. nov., comprising a distinct lineage of the Archaea. Int. J. Syst. Evol. Microbiol. 2000;50:997–1006. doi: 10.1099/00207713-50-3-997. [DOI] [PubMed] [Google Scholar]
- 73.Schleper C., Puehler G., Holz I., Gambacorta A., Janekovic D., Santarius U., et al. Picrophilus gen. nov., fam. nov.: a novel aerobic, heterotrophic, thermoacidophilic genus and family comprising archaea capable of growth around pH 0. J. Bacteriol. 1995;177:7050–7059. doi: 10.1128/jb.177.24.7050-7059.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kurosawa N., Itoh Y.H., Iwai T., Sugai A., Uda I., Kimura N., et al. Sulfurisphaera ohwakuensis gen. nov., sp. nov., a novel extremely thermophilic acidophile of the order Sulfolobales. Int. J. Syst. Evol. Microbiol. 1998;48:451–456. doi: 10.1099/00207713-48-2-451. [DOI] [PubMed] [Google Scholar]
- 75.Guzman L.M., Belin D., Carson M.J., Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stanley N.R., Palmer T., Berks B.C. The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J. Biol. Chem. 2000;275:11591–11596. doi: 10.1074/jbc.275.16.11591. [DOI] [PubMed] [Google Scholar]
- 77.Sargent F., Stanley N.R., Berks B.C., Palmer T. Sec-independent protein translocation in Escherichia coli. J. Biol. Chem. 1999;274:36073–36082. doi: 10.1074/jbc.274.51.36073. [DOI] [PubMed] [Google Scholar]
- 78.Kneuper H., Maldonado B., Jäger F., Krehenbrink M., Buchanan G., Keller R., et al. Molecular dissection of TatC defines critical regions essential for protein transport and a TatB–TatC contact site. Mol. Microbiol. 2012;85:945–961. doi: 10.1111/j.1365-2958.2012.08151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.