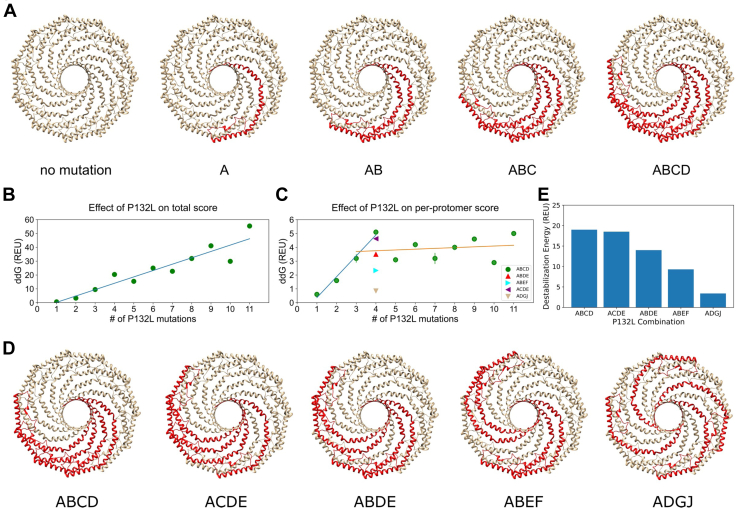
Figure 5.
Computational predictions of the energetic cost of incorporating P132L protomers into mixed complexes.A, schematic of CAV1 complexes containing increasing numbers of P132L mutant protomers. In this scenario, mutant protomers are located directly adjacent to one another. B, Rosetta ddG for CAV1 complexes containing increasing numbers of P132L mutants located directly adjacent to one another as in panel A. C, as in B, except here, Rosetta ddG was normalized per number of P132L mutations. Lines are drawn to guide the eye to highlight differences in predicted stability when one to three copies of P132L (blue line) versus four or more (orange line) positioned directly adjacent to one another are present in a mixed complex with WT CAV1 (green circles). For comparison, per-protomer Rosetta ddG values are shown for mixed complexes containing four copies of P132L arranged at different spacings (colored triangles) as shown in panel D. D, schematic of CAV1 complexes containing four copies of P132L arranged at different spacings. E, Rosetta ddG values for CAV1 complexes containing four P132L protomers arranged at different spacings shown in panel D. REU, Rosetta energy units.