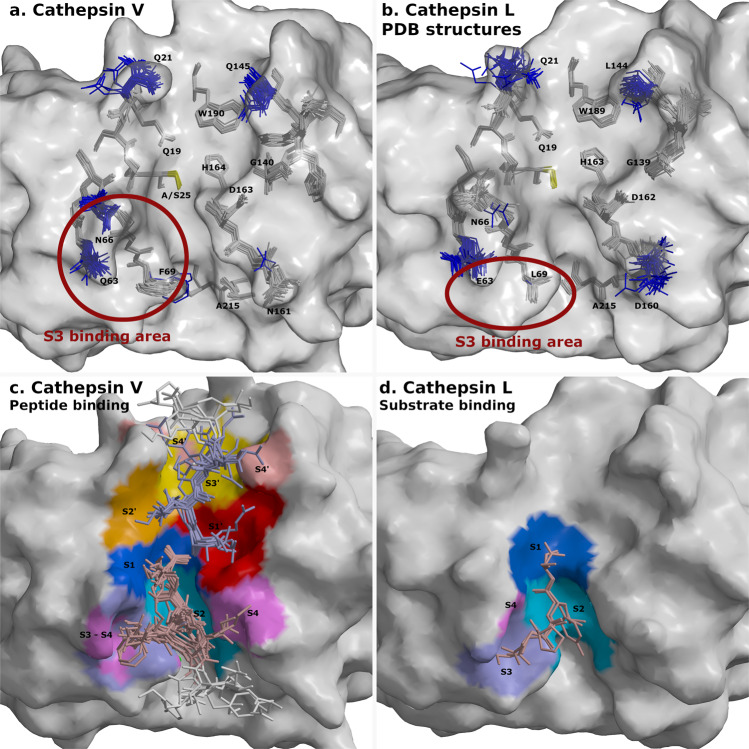
Fig. 7. Flexible and rigid residues of cathepsins V and L and their substrate-binding areas.
Surfaces of cathepsins V and L are shown in gray. Catalytic residues at site 25 are shown in yellow. Peptides and cathepsin residues are presented as sticks. a Superimposed structures of cathepsin V-peptide complexes. Flexible cathepsin V residues that provided versatile binding area for peptide binding are shown in blue. Rigid residues are shown in gray. b Superimposed structures of cathepsins L from PDB database with the equivalent labeling (PDB entries 1CJL, 1CS8, 1ICF, 1MHW, 2NQD, 2XU1, 2XU3, 2XU4, 2XU5, 2YJ2, 2YJ8, 2YJ9, 2YJB, 3BC3, 3H89, 3H8B, 3H8C, 3HHA, 3HWN, 3K24, 3KSE, 3OF8, 3OF9, 4AXL, 4AXM, 5F02, 5MAE, 5MQY, 6EZP, 6EZX, 6F06, 6JD0, and 6JD8). The structure of C25A mutant with SO42- ion in the active site (3IV2) is not included due to distorted active site. Red circles in panels a and b depict S3 binding area of both cathepsins. c Binding areas of peptides at positions from P4-P4′ are shown in color spectra from blue to magenta at the non-primed side and from red to rose at the primed side. Peptide residues from P1-P4 and P1′-P4′ are shown in pale pink and pale blue, respectively, whereas the residues beyond P4 and P4′ are in white. d Processed peptide and protein substrates of cathepsin L structures (3K24, 5I4H) at the non-primed side. Their binding areas are presented with the same coloring annotation as in panel c. The figures were generated with MAIN59 and rendered with RASTER 3D66.