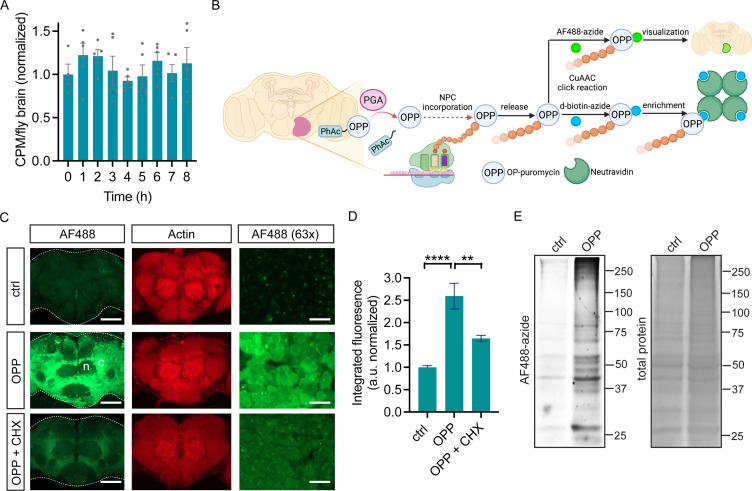
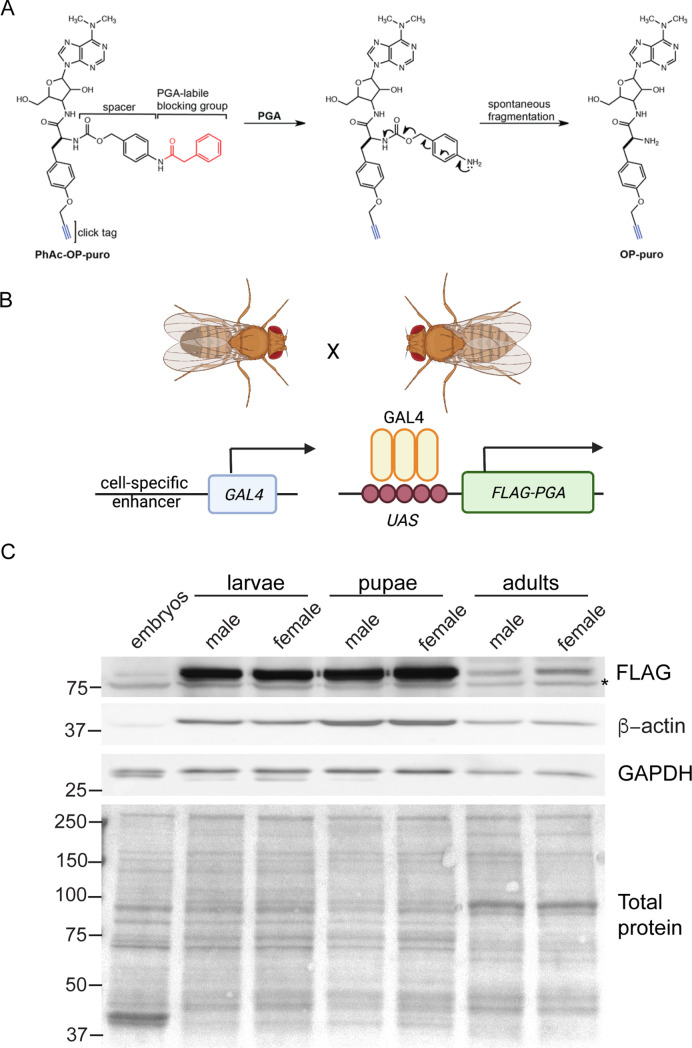
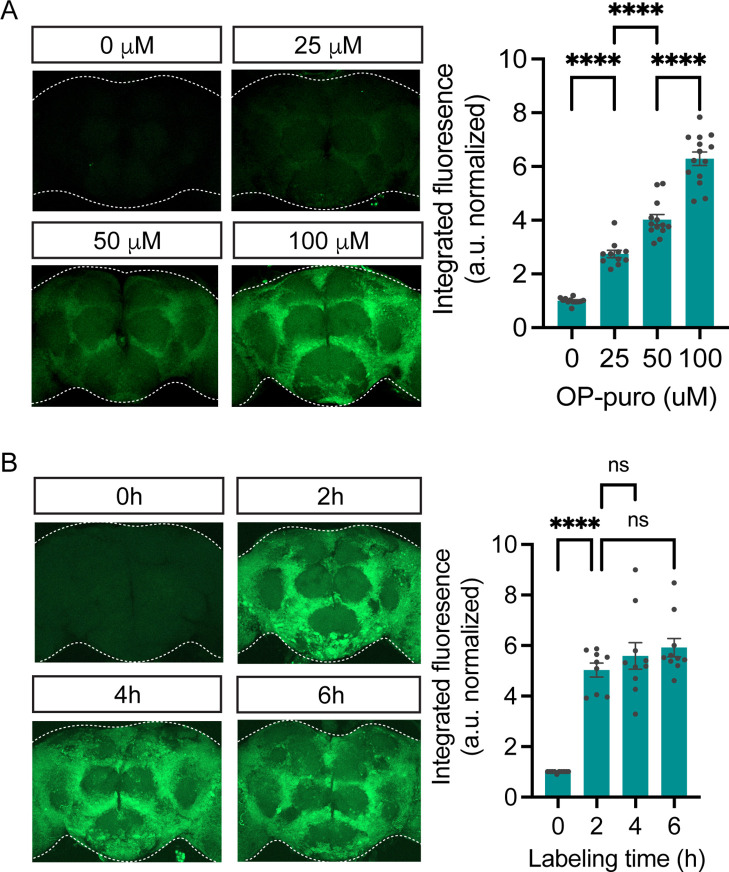
Figure 1. O-propargyl-puromycin (OPP) labeling in Drosophila brain.
(A) Global protein synthesis (35S-met/cys incorporation) is unaltered for 8 hr in newly-isolated w1118 whole brain preparations (ANOVA, n.s. n=4–5 groups of 8–10 brains per timepoint). (B) Schematic illustrating cell type-specific protein synthesis labeling by PGA-dependent OPP incorporation (POPPi) for visualization or capture of the nascent proteome. Spatially targeted penicillin G acylase (PGA) expression catalyzes phenylacetyl-OPP (PhAc-OPP) blocking group removal, liberating OPP for incorporation into nascent polypeptide chains (NPC). OP-puromycylated proteins can be visualized by confocal microscopy following conjugation to a fluorescent-azide or enriched following conjugation to desthiobiotin-azide. (C) Newly-synthesized protein (from w1118 brains) visualized by AF488-azide after OPP incubation but not without OPP (ctrl) and diminished signal with cycloheximide (CHX). Labeling appears stronger in cell bodies within the cell cortex (C) than in the neuropil (n). Higher magnification (63 x) images are from the cell cortex. Scale bars are 60 μM (or 10 μM for 63 x). (D) Quantitation of (C) revealing a significant effect of the treatment group (ANOVA, p<0.0001, Bonferroni post-test, **p<0.01, ****p<0.0001, n=8–11 brains/group). (E) In-gel fluorescence of brain protein extracts, ctrl is no OPP. Data are mean ± SEM. Schematic in B created on Biorender. See also Figure 1—figure supplements 1 and 2.