Figure 1. DUBTAC platform.
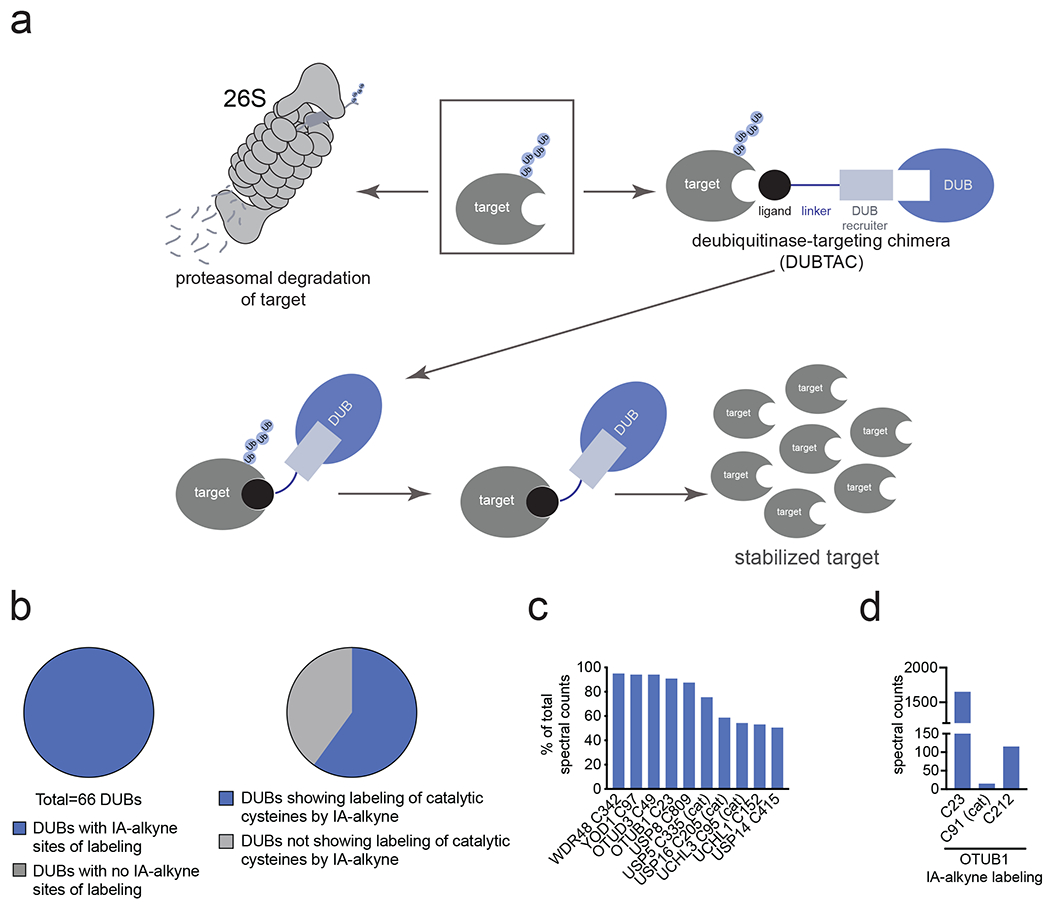
(a) DUBTACs are heterobifunctional molecules consisting of a protein-targeting ligand linked to a DUB recruiter via a linker. DUBTACs are ideally used for stabilizing the levels of actively ubiquitinated proteins that are degraded by the proteasome. When treated in cells, a DUBTAC will induce the proximity of a DUB with a target protein to remove polyubiquitin chains to prevent the protein from undergoing proteasome-mediated degradation, thereby stabilizing and elevating the level of the actively degraded protein. (b) Out of 65 DUBs mined in our research group’s aggregate chemoproteomic datasets of cysteine-reactive probe labeling with IA-alkyne in various complex proteomes, we identified probe-modified cysteines across all 100 % of the 65 DUBs. This is shown in the first pie chart. Among the 65 DUBs that showed probe-modified cysteines, 40 of these DUBs showed >10 aggregate spectral counts across our chemoproteomic datasets. 24 DUBs, or 60 %, of these 40 DUBs showed labeling of the DUB catalytic or active site cysteines. (c) Mining the DUB data, we identified 10 DUBs wherein there was one probe-modified cysteine that represented >50 % of the total aggregate spectral counts for probe-modified cysteine peptides for the particular DUB. 7 of those 10 DUBs do not target a known catalytic cysteine whereas 3 do target the catalytic cysteine (abbreviated by cat). (d) Analysis of aggregate chemoproteomic data for OTUB1 IA-alkyne labeling showing that C23 is the dominant site labeled by IA-alkyne compared to the catalytic (cat) C91. Chemoproteomic data analysis of DUBs across aggregated datasets can be found in Table S1.
