Extended Data Figure 2. NMR analysis of OTUB1, EN523, and UBE2D2.
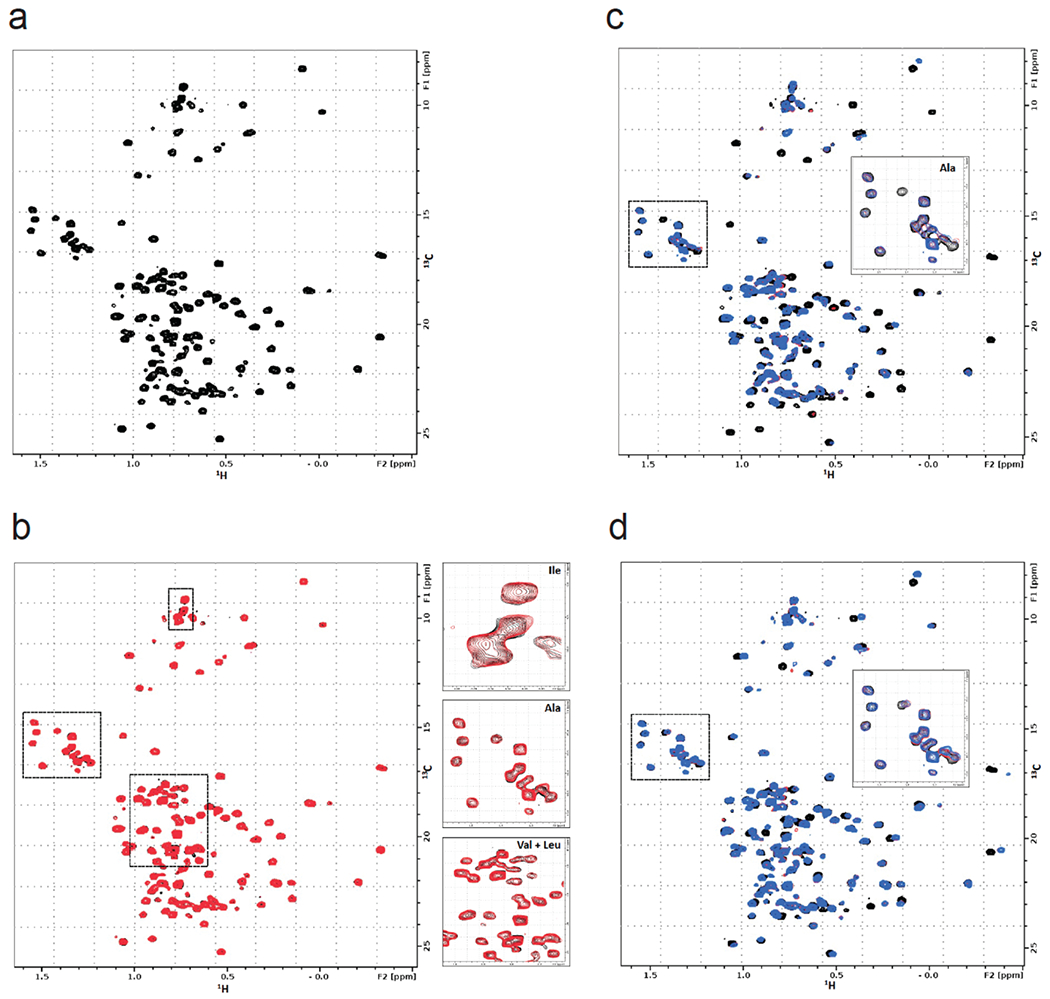
(a) 13C-HMQC spectrum of OTUB1 labeled on methyl groups of isoleucine, alanine, valine and leucine residues. The presence of peaks with negative proton chemical shifts indicates that the protein is properly folded. (b) Overlay of HMQC spectra of apo-OTUB1 (black) and EN523-bound OTUB1 (red). While both spectra are mostly identical, we identified small but clear chemical shift perturbations of alanine, isoleucine, valine and leucine peaks. Some of these signal changes are shown in the respective blow-up boxes. (c) Overlay of HMQC spectra of apo-OTUB1 (black), UBE2D2 bound OTUB1 (red) and EN523/UBE2D2-bound OTUB1 (blue). The strong chemical shift perturbations (CSPs) are evidence of specific interactions between OTUB1 and the ubiquitinylated ubiquitin-conjugating enzyme. The lack of significant differences between spectra recorded in the presence and absence of EN523 prove that the covalent ligand does not interfere with the protein-protein interaction. Differing peak shift pattern are only seen for peaks directly affected by compound binding (see inlay for blow-up of Ala region). (d) Overlay of HMQC spectra of apo-OTUB1 (black), Ub-UBE2D2 bound OTUB1 (red) and EN523/Ub-UBE2D2-bound OTUB1 (blue). The strong CSPs are evidence of specific interactions between OTUB1 and the ubiquitin-conjugating enzyme. The lack of significant differences between spectra recorded in the presence and absence of EN523 prove that the covalent ligand does not interfere with the protein-protein interaction. Differing peak shift pattern are only seen for peaks directly affected by compound binding (see inlay for blow-up of Ala region).
