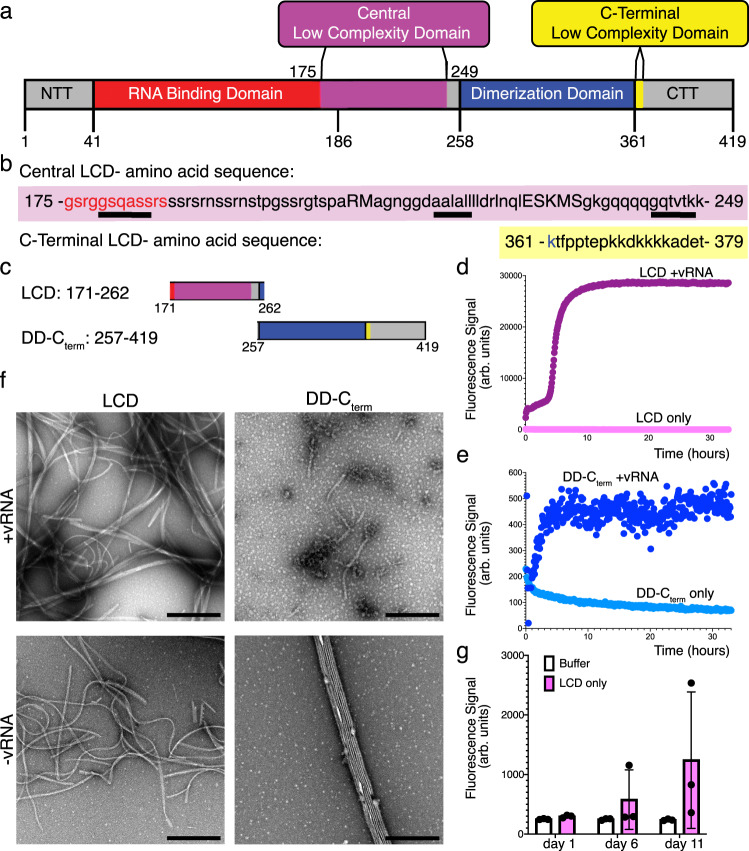
Fig. 1. NCAP’s LCDs form fibrils and ThT-positive species.
a NCAP’s domain organization. Domain definitions: N-terminal tail (NTT, gray), RNA-binding domain (red); Central low complexity domain (LCD, purple; residues 175–249), Dimerization domain (blue); C-terminal tail (CTT, gray). The C-terminal LCD is highlighted in yellow (residues 361–379). b Amino acid sequence of the central and C-terminal LCDs highlighted and colored according to the color scheme in (a). Lowercase letters represent residues of low complexity while capital letters represent non-low-complexity residues. No more than five interrupting non-low-complexity residues between strings of 10 or more low-complexity residues were allowed. Steric-zipper forming sequences that are discussed below are underlined in the sequence of the central LCD. c Protein segments used in this study are abbreviated as LCD, consisting of the central LCD and surrounding residues, and as DD-Cterm, consisting of the dimerization domain (DD) and the C-terminal tail, including the C-terminal LCD. The LCD and DD-Cterm segments are colored according to the color scheme in (a). d and e ThT fibril formation kinetic assays of the LCD (d) and DD-Cterm (e) segments incubated with (purple/navy, respectively) and without (pink/light blue, respectively) hairpin-Site2 (S2hp) viral RNA (vRNA). f Fibril formation from concentrated LCD and DD-Cterm samples observed by negative stain EM after 6 days of incubation with and without S2hp vRNA. Scale bar = 500 nm. g Endpoint ThT fluorescence measurements of concentrated LCD-only samples (pink) and buffer-only controls (white) at days 1, 6, and 11 of incubation. Dots indicate individual data points and bars represent mean values ± SD. n = 3 samples. Source data for panels d, e, and g are provided as a Source Data file.