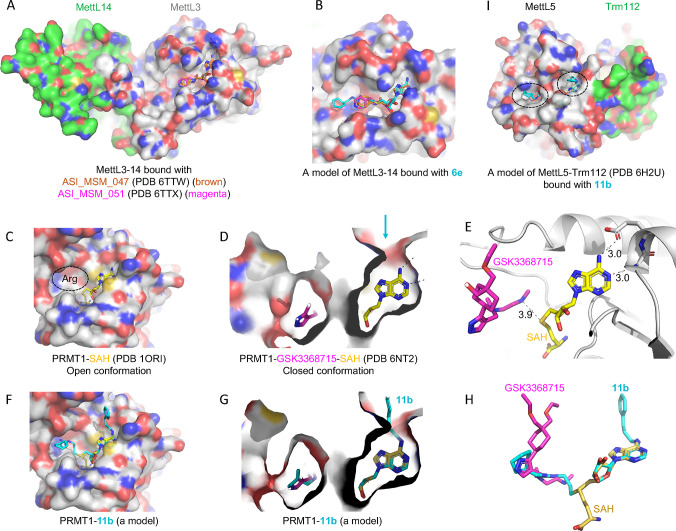
Figure 5.
Models of compounds 6e or 11b in binding MettL3, PRMT1, and MettL5. (A) Structures of MettL3–MettL14 bound with two inhibitors (PDB 6TTW and PDB 6TTX). (B) A model of 6e binding MettL3 by superimposing the adenosine moiety. (C) Structure of PRMT1–SAH (PDB 1ORI) where the SAH binding site is exposed while the N-terminal residues are disordered in the absence of bound arginine substrate (circled). (D) Structure of PRMT1–SAH–GSK3368715 (PDB 6NT2) where SAH is largely buried in the presence of substrate-competitor inhibitor. The arrow indicates a narrow opening near the adenine N6-amino group. (E) The distance is 3.9 Å between the sulfur atom of SAH and the GSK inhibitor which occupied the substrate arginine binding site. (F) A model of compound 11b binding PRMT1 in the open conformation. (G, H) A model of compound 11b binding PRMT1 in the closed conformation. Compound 6e could be modeled the same way without the N6-addition. (I) Structure of MettL5–Trm112 in complex with SAM (PDB 6H2U). A model of 11e binding MettL5 by superimposing the adenosine moiety, and the aliphatic and aromatic rings (circled) are visible from the surface.