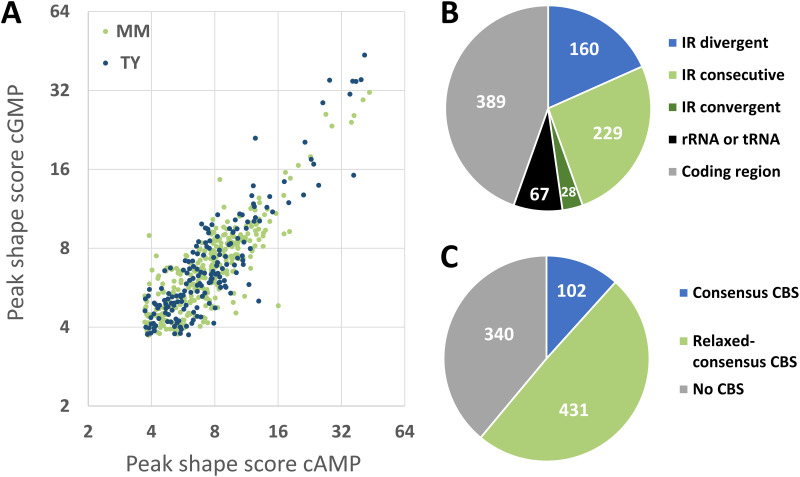
FIG 1.
ChIP-seq-assisted screening for Clr-cAMP and Clr-cGMP binding sites. (A) Correlation between peak shape score values of ChIP-seq peaks detected in samples of cells grown in TY or MM medium, both supplemented with either cAMP or cGMP. (B) Location of Clr ChIP-seq peaks within genomic features. IR, intergenic region; IR divergent, IR between two genes transcribed in divergent directions; IR consecutive, IR between two genes transcribed in the same direction; IR convergent, IR between two genes transcribed in converging directions; rRNA or tRNA, rRNA or tRNA encoding region; coding region, protein coding region. (C) Presence of putative Clr binding sites (CBSs) in the vicinity of ChIP-seq peaks. Consensus CBS, GTNNCNNNNGNNAC. Relaxed-consensus CBS, one mismatch with the consensus was allowed.