ABSTRACT
DNA-damage tolerance (DDT) is a mechanism by which eukaryotes bypass replication-blocking lesions to resume DNA synthesis and maintain cell viability. In Saccharomyces cerevisiae, DDT is mediated by sequential ubiquitination and sumoylation of proliferating cell nuclear antigen (PCNA, encoded by POL30) at the K164 residue. Deletion of RAD5 or RAD18, encoding two ubiquitin ligases required for PCNA ubiquitination, results in severe DNA-damage sensitivity, which can be rescued by inactivation of SRS2 encoding a DNA helicase that inhibits undesired homologous recombination. In this study, we isolated DNA-damage resistant mutants from rad5Δ cells and found that one of them contained a pol30-A171D mutation, which could rescue both rad5Δ and rad18Δ DNA-damage sensitivity in a srs2-dependent and PCNA sumoylation-independent manner. Pol30-A171D abolished physical interaction with Srs2 but not another PCNA-interacting protein Rad30; however, Pol30-A171 is not located in the PCNA-Srs2 interface. The PCNA-Srs2 structure was analyzed to design and create mutations in the complex interface, one of which, pol30-I128A, resulted in phenotypes reminiscent of pol30-A171D. This study allows us to conclude that, unlike other PCNA-binding proteins, Srs2 interacts with PCNA through a partially conserved motif, and the interaction can be strengthened by PCNA sumoylation, which turns Srs2 recruitment into a regulated process.
KEYWORDS: Saccharomyces cerevisiae, PCNA, DNA-damage tolerance, Rad5, Rad18, Srs2, PIP box, budding yeast
INTRODUCTION
Living organisms are constantly exposed to spontaneous and environmental DNA damage. In addition to various DNA repair pathways, cells have also evolved to survive in the presence of replication-blocking lesions, a process known as DNA-damage tolerance (DDT). In a model lower eukaryote Saccharomyces cerevisiae, DDT is achieved by sequential ubiquitination of proliferating cell nuclear antigen (PCNA) (1, 2). In response to DNA damage, an E2-E3 complex Rad6-Rad18 monoubiquitinates PCNA at its K164 residue, which increases affinity for Y-family DNA polymerases, including Polη (3) and Rev1 (4), and promotes translesion DNA synthesis (TLS). Monoubiquitinated PCNA can be further polyubiquitinated through K63 linkage by another E2-E3 complex Mms2-Ubc13-Rad5 that leads to error-free lesion bypass (5), probably through template switch (6, 7). The same K164 residue in PCNA can also be sumoylated by an E2-E3 complex Ubc9-Siz1 in the absence of DNA damage (5), which recruits Srs2 to inhibit undesired homologous recombination (HR) (8, 9). The srs2 (suppression of rad-six) mutant was initially isolated by its ability to rescue the severe DNA-damage sensitivity of rad6 and rad18 mutants (10) and subsequently found to encode a DNA helicase (11) that inhibits hyper-recombination (12) by preventing Rad51-ssDNA filament formation (13, 14). Srs2 also suppresses the severe DNA-damage sensitivity of other DDT pathway mutants, including rad5 (15–17). Since the srs2 suppression of DNA-damage sensitivity is limited to only DDT pathway mutants and acts to channel DNA lesions to HR (18), loss of Srs2 is thought to activate a salvage HR pathway (19, 20).
PCNA forms a toroidal-shaped homotrimer that interacts with the DNA strand by encircling it, forming a sliding clamp that recruits a large number of proteins to the DNA replication fork not only for replication, but also for other processes, including cell cycle regulation, recombination and DNA damage response (DDR) (21). The most common PCNA-binding motif is known as the PCNA interaction protein (PIP) box, consisting of a consensus sequence Q-x-x-(h)-x-x-(a)-(a) (where “h” represents amino acid with moderately hydrophobic side chains like L, I, M; “a” represents amino acid with highly hydrophobic, aromatic side chains like F, Y; and “x” represents any residues) (22). Several proteins involved in the yeast DDT pathway, including Srs2 (23), Polη (encoded by RAD30) (24, 25), Pol2 and all three Polδ subunits (26), contain a PIP box or its variants. Correspondingly, analysis of the PCNA-PIP box complex structure (27) reveals that PCNA interdomain connecting loop (IDCL) and C-terminal regions mediate its interaction with the PIP box.
We systematically analyzed DNA-damage resistant isolates from rad5 and rad18 null mutants. While majority of these isolates contained mutations in the SRS2 gene as anticipated, several isolates from rad5Δ cells did not contain srs2 mutation. Whole-genome sequencing analyses identified putative mutations responsible for DNA-damage tolerance phenotype, among which a missense mutation in POL30 encoding yeast PCNA is further investigated in this study. It was found that PCNA mutations specifically affecting its interaction with Srs2 can activate the salvage HR pathway.
RESULTS
Isolation and characterization of MMS-resistant colonies from rad5Δ and rad18Δ cells.
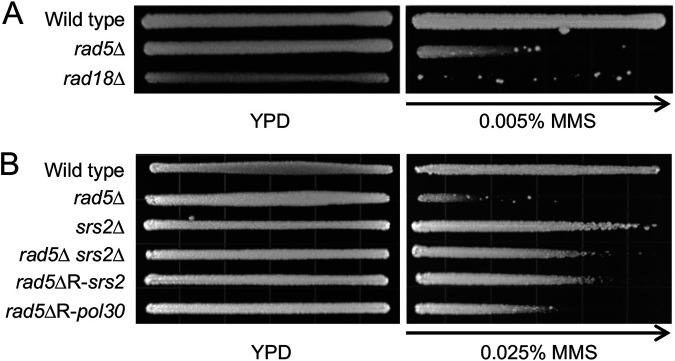
Rad18 and Rad5 serve as E3s for PCNA monoubiquitination and subsequent polyubiquitination, respectively (5). In addition, Rad5 is also required for the recruitment of a TLS polymerase Rev1 (28). Hence, Rad18 and Rad5 are required for both branches of DDT, and their null mutants are extremely sensitive to DNA-damaging agents that block replication, including ultraviolet (UV) irradiation, methyl methanesulfonate (MMS), and 4-nitroquinoline N-oxide (4-NQO). It was routinely observed that in an MMS gradient plate assay, some resistant colonies appeared at the high MMS concentration end (Fig. 1A). These resistant colonies turned out to be inherited, as they became significantly more resistant than their parental cells when replating on the MMS gradient plates again (see Fig. 1B for example). It has been well known that srs2 loss-of-function mutations can rescue rad6, rad18 (10) and rad5 (15–17) severe sensitivity. To ask if these mutants carry srs2 mutations, we sequenced the SRS2 and its promoter region from 33 independent MMS-resistant rad18Δ isolates and found that they all carried mutations in SRS2 (data not shown). Interestingly, out of 25 independent MMS-resistant rad5Δ isolates (designated rad5ΔR), only half (13/25) carried srs2 mutations. Genomic DNA was extracted from the 13 non-SRS2-mutation rad5ΔR isolates, and the whole-genome sequencing was performed and analyzed. Indeed, these isolates did not contain mutation in SRS2, and the bioinformatics analysis revealed candidate gene mutations, among which a pol30-A171D mutation caught our attention. Fig. 1B shows that a sample rad5ΔR isolate that contains a srs2 mutation (rad5ΔR-srs2) is indistinguishable from the reconstituted rad5Δ srs2Δ mutant. In comparison, a rad5ΔR isolate containing a pol30-A171D (designated rad5ΔR-pol30) mutation is much more resistant than rad5Δ but less resistant than the rad5Δ srs2Δ mutant.
FIG 1.
rad5Δ, rad18Δ and their MMS resistant mutant phenotypes by an MMS gradient plate assay. (A) rad5Δ and rad18Δ mutant phenotypes. (B) Relative sensitivity of MMS resistant isolates (rad5ΔR) from rad5Δ cells. Cells grown on a YPD plate serve as controls. The length of growth on the MMS-containing gradient plates measures the relative sensitivity of cells to MMS. Arrows point to increasing MMS concentration. Plates were incubated at 30°C for 2 days before photography.
pol30-A171D is synergistic to pol30-K164R in the rescue of rad5Δ and rad18Δ sensitivity.
To ask whether the pol30-A171D mutation is indeed responsible for the rescue of rad5Δ cells, we wished to create a pol30-A171D rad5Δ double mutant. However, POL30 is an essential gene that cannot be readily deleted and replaced by a point mutation. Hence, we took a plasmid shuffling approach as described (29), in which the chromosomal POL30 was deleted while carrying a YCp-Pol30 plasmid with a URA3 selectable marker. Another plasmid carrying a YCp-Pol30 plasmid or its mutant derivatives with a LEU2 selectable marker can be transformed into the above cells and the transformants lost original YCpU-Pol30 can be selected in a 5-FOA plate. The phenotype of a pol30 mutant can then be assessed in comparison to its wild type transformant.
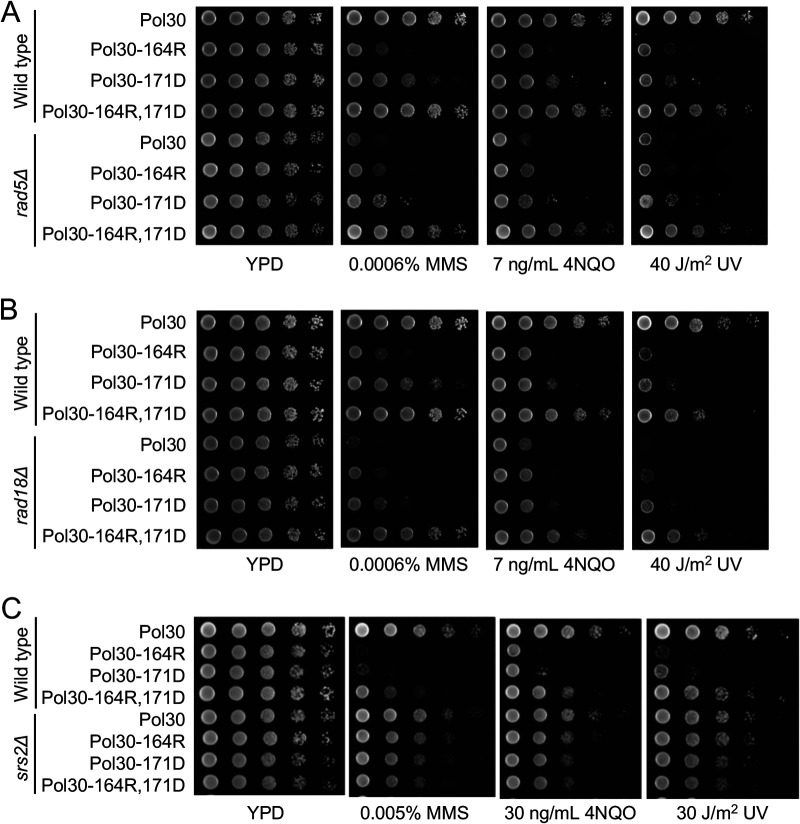
The reconstituted pol30-A171D mutant displayed increased sensitivity to MMS, and it was able to rescue the severe MMS sensitivity of the rad5Δ mutant (Fig. 2A). The pol30-K164R mutation was reported to rescue the rad18Δ sensitivity (5). It was observed that in comparison to pol30-K164R, pol30-A171D cells were less sensitive to DNA-damaging agents both in wild-type and rad5Δ backgrounds (Fig. 2A). In addition, pol30-A171D also rescued the rad18Δ mutants to DNA damage (Fig. 2B).
FIG 2.
Genetic interactions between pol30-A171D and relevant mutations by a serial dilution assay. (A) Genetic interaction between pol30-A171D and pol30-K164R in the rescue of rad5Δ mutant. (B) Genetic interaction between pol30-A171D and pol30-K164R in the rescue of rad18Δ mutant. (C) Genetic interaction between pol30-A171D and pol30-K164R in the srs2 background. Yeast cells cultured overnight were used to make 10-fold serial dilutions and then spotted to YPD or YPD plus various concentrations of MMS or 4NQO or exposed to UV irradiation at indicated doses, followed by incubation at 30°C for 2 days before photography. Only one representative dose for each treatment is shown.
Since Pol30-K164 is the ubiquitination and sumoylation site (5), it is believed that the pol30-K164R mutation rescues rad18 sensitivity through loss of PCNA sumoylation and compromised Srs2 recruitment (8, 9). We first asked whether pol30-A171D affects PCNA sumoylation by a Western blot analysis, which revealed that under the same experimental conditions, pol30-K164R abolished sumoylation at the Pol30-K164 residue while pol30-A171D did not (Supplementary Fig, S1, lanes 2 and 4). To address genetic relationship between pol30-K164R and pol30-A171D, we created and tested the pol30- K164R,A171D double mutant. To our surprise, the double mutant was much more resistant to all three DNA-damaging agents than its corresponding single mutant, and fully rescued rad5Δ (Fig. 2A) and rad18Δ (Fig. 2B) mutants. In quantitative terms, the pol30-K164R mutation rescued rad5Δ or rad18Δ MMS sensitivity by less than 10-fold, and the pol30-A171D mutation rescued rad5Δ or rad18Δ MMS sensitivity by more than 10-fold, while the double mutation rescued their MMS sensitivity by as high as 10,000-fold. Hence, pol30-K164R and pol30-A171D are synergistic in rescuing rad5Δ and rad18Δ MMS, as well as 4NQO and UV sensitivity.
The rescuing effect of pol30-A171D relies on SRS2.
The rescuing effect of pol30-K164R is known to rely on SRS2. To ask whether pol30-A171D also behaves the same, we tested the relative sensitivity of pol30-A171D, pol30-K164R and the corresponding double mutant in a srs2 background. As shown in Fig. 2C, the srs2 mutation is epistatic to all three pol30 mutations with respect to DNA damage sensitivity. The above observations allow us to conclude that pol30-A171D acts in an SRS2-dependent manner.
pol30-A171D affects Pol30-Srs2 interaction.
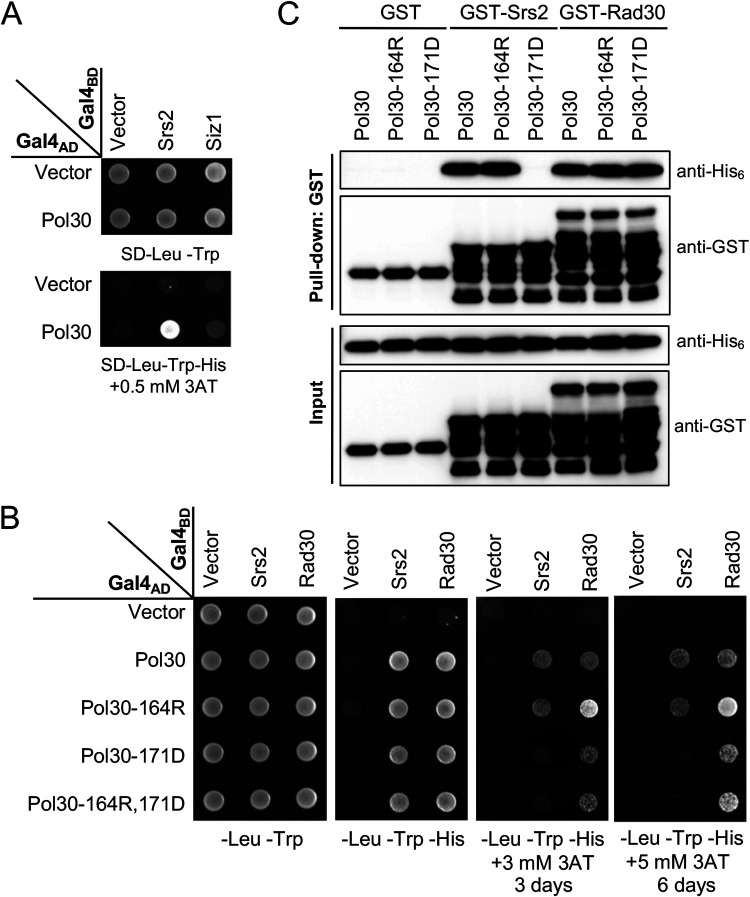
Through the above genetic analysis, we hypothesized that the pol30-A171D mutation affects Srs2 recruitment independently of PCNA sumoylation. To test this hypothesis, A yeast two-hybrid (Y2H) assay was performed to examine physical interaction between Pol30 and Srs2 as well as Siz1. Under our experimental conditions, Pol30 was found to interact with Srs2 but not Siz1 (Fig. 3A). Srs2 was then used to test its interaction with Pol30 mutations using a PIP box-containing protein Rad30 as a positive control. Fig. 3B shows that Pol30-A171D did not affect interaction with Rad30 but severely reduced interaction with Srs2. In contrast, Pol30-K164R did not affect interaction with Srs2 and even enhanced interaction with Rad30. Again, the dual substitution behaved like Pol30-A171D.
FIG 3.
Physical interaction between Pol30-A171D and Srs2. (A) Interaction between Pol30, Srs2 and Siz1 by a Y2H assay. (B) Interaction between mutated Pol30 and Srs2, Rad30 by a Y2H assay. SRS2, SIZ1 and RAD30 ORFs were cloned in pGBT9 (Gal4BD), and POL30 ORF and its mutant alleles were cloned in pGAD424 (Gal4AD). pGBT9 and pGAD424 derived plasmids were cotransformed into PJ69-4a, and the transformants were spotted on control (SD-Leu-Trp) and selective (SD-Leu-Trp-His with or without 3AT) plates, which were incubated at 30°C for 3 days or as indicated before photography. Independent transformants were examined in parallel on multiple selective plates and incubated for different periods. Only the representative images are shown. (C) In vitro interaction between Srs2 C terminus (Srs2-CT), Rad30-PIP and Pol30 or its mutant derivatives by a GST pulldown assay. Proteins before and after the GST pulldown were subjected to Western blotting by using antibodies against His6 and GST tags.
The Srs2 C-terminal domain (Srs2-CT) was reported to interact with Pol30 and SUMO through its PIP box and SIM motif, respectively (23). In a GST-pulldown assay, Srs2-CT (amino acid residues 1107 to 1174) was cloned into pGEX6 to produce GST-tagged Srs2-CT and His6-tagged Pol30 was produced by pET-Pol30 transformed bacterial cells. Under a robust condition of direct interaction between GST-Srs2-CT and His6-Pol30, Pol30-K164R did not affect this interaction while Pol30-A171D completely abolished such interaction; in contrast, neither Pol30-K164R nor Pol30-A171D affected physical interaction with the Rad30-PIP domain (Fig. 3C). In conclusion, both Y2H and GST-pulldown assay results support a notion that Pol30-A171D affects Srs2 interaction, but Pol30-K164R does not. It is noted that in both Y2H and pulldown assays, Pol30 was not sumoylated, which explains why Pol30-K164R does not affect interaction with Srs2.
Distinct effects between pol30-A171D and pol30-K127R.
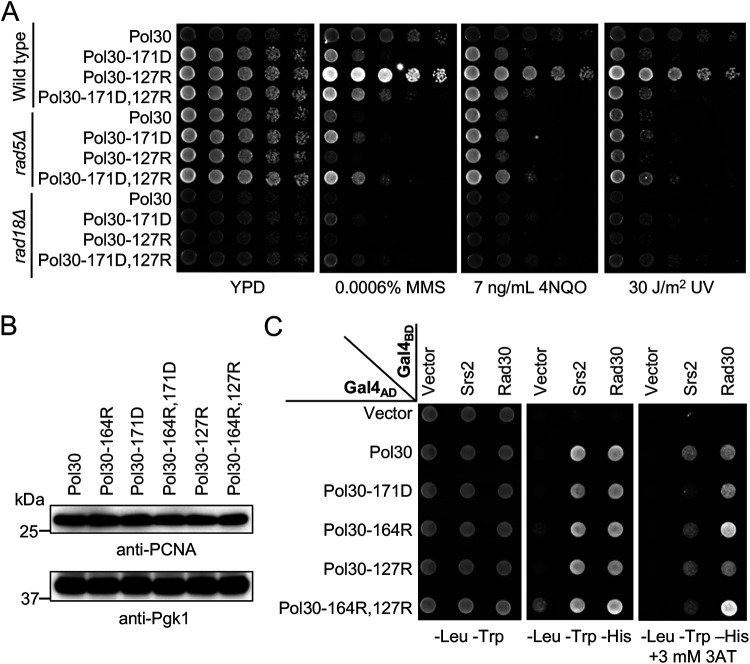
Pol30-K127 can also be sumoylated (5), which regulates PIP-box proteins like EcoI for the sister chromatid cohesion (30). It was previously reported that pol30-K127R and pol30-164R mutations are synergistic in rescuing the rad18 severe DNA-damage sensitivity (8, 9, 31), implying that sumoylation at the Pol30-K127 residue can backup Pol30-K164 sumoylation for the Srs2 recruitment. It raises a possibility that the Pol30-A171D substitution interferes with Pol30-K127 sumoylation, resulting in the observed mutant phenotypes. Indeed, under our experimental conditions, pol30-K127R itself does not display altered DNA-damage sensitivity or rescuing effects on rad5 and rad18 but causes synergistic phenotypes when combined with pol30-K164R (Fig. S2), reminiscent of the pol30-A171D,K164R double mutant (Fig. 2A,B). However, pol30-A171D and pol30-K127R are additive in DNA-damage response (Fig. 4A), indicating that they function in different pathways. The observed phenotypes were not due to altered cellular PCNA levels, as pol30-A171D, pol30-K164R, pol30-K127R and their corresponding double mutations do not affect protein stability (Fig. 4B). Furthermore, while Pol30-A171D fails to bind Srs2, Pol30-K127R and Pol30-K127R,K164R substitutions do not affect Srs2 interaction (Fig. 4C), further confirming that pol30-A171D rescues rad5 and rad18 independently of Pol30-K127 sumoylation.
FIG 4.
Functional characterization of the pol30-K127R mutation. (A) Genetic interaction between pol30-K127R and pol30-A171D. Experimental conditions are as described in Fig. 2. (B) Western blot analysis of cellular PCNA levels. Pol30 variants are as indicated on the top panel. Pgk1 was used as a loading control. (C) Physical interaction between Srs2 and Pol30 amino acid substitutions by a Y2H assay. Experimental conditions are as described in Fig. 3B, and the plates were incubated at 30°C for 3 days before photography.
Genetic interactions between pol30-K164R and pol30-K127R in rescuing severe DNA-damage sensitivities of rad5 and rad18 mutants by a serial dilution assay. Download FIG S2, DOCX file, 0.2 MB (217.9KB, docx) .
Copyright © 2023 Fan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Assessment of PCNA IDCL and C-terminal mutations.
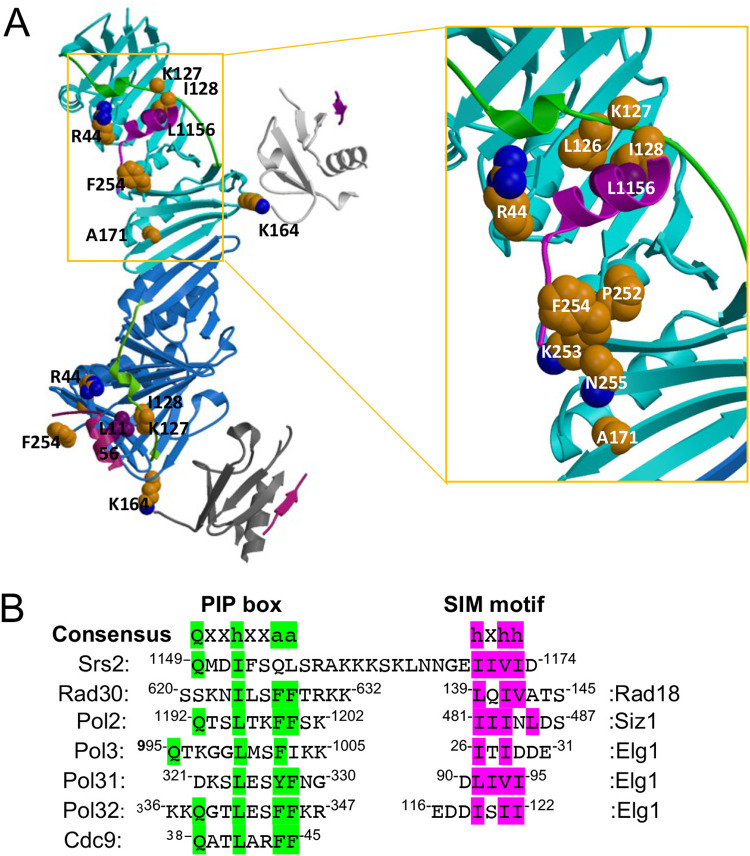
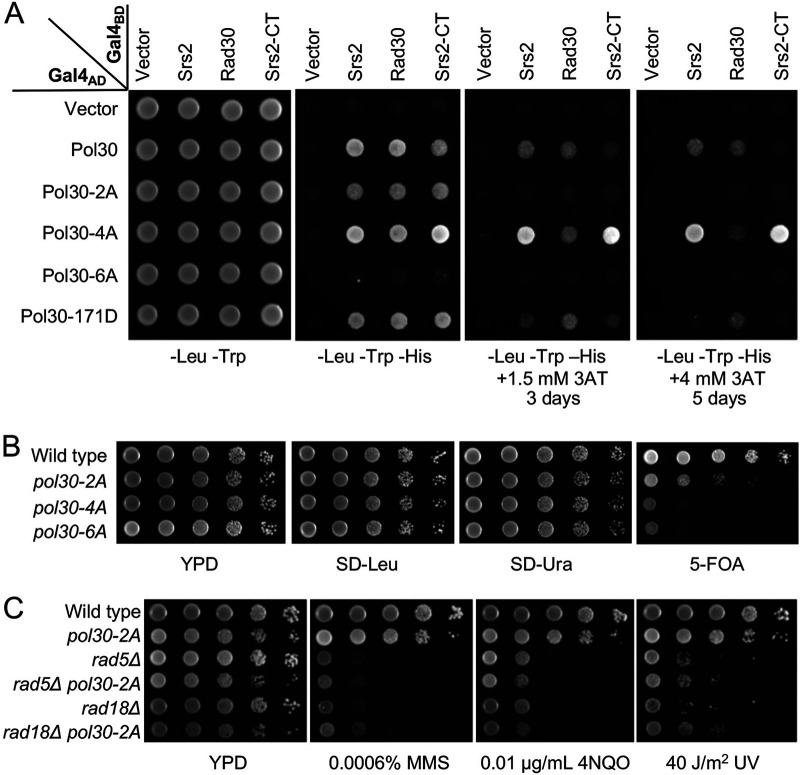
To ask how the Pol30-A171D substitution affect its interaction with Srs2, we looked at the reported SUMO-PCNA-Srs2-CT complex structure (23). To our surprise, Pol30-A171 is not located in the PCNA-Srs2 interface (Fig. 5A). Srs2-CT has been reported to contain an “atypical PIP box” (23). Comparison of different sources of PIP-box sequences (Fig. 5B) revealed that among conserved residues, Srs2-CT contains an N-terminal “Qxxh” sequence, but not the C-terminal “FF” sequence. In contrast, some other proteins, including Rad30 contain the C-terminal “hxxFF” sequence but not the N-terminal “Q” residue. Since PIP box has been reported to bind two separate regions in PCNA, namely, IDCL (residues 121 to 132) and the C terminus (residues 251 to 258) (27), it is hypothesized that mutations in one of the two PCNA regions result in reduced PCNA-Srs2 interaction and the rescue of rad5Δ/rad18Δ mutant phenotypes. To test this hypothesis, two site-specific mutations were created: pol30-2A (L126A, I128A) was to affect the IDCL region and pol30-4A (252-255AAAA) was to affect the C-terminal region (Fig. 5A). In addition, pol30-6A (2A + 4A) was the combination of the above two mutations (Fig. 5A).
FIG 5.
Protein structure and sequence analyses of the SUMO-PCNA-Srs2-CT complex. (A) The SUMO-PCNA-Srs2-CT complex structure from Protein Data Bank 3V62. Left panel: Two subunits of Pol30 (cyan and blue), SUMO (light and dark gray) and Srs2-CT (magenta, the linker between the PIP-like box and SIM is disordered) are shown. The two IDCL domains are highlighted in green. Right panel: an enlarged view from the boxed region on the left. Sidechains of discussed residues are shown in space-filling models with carbon atoms colored in gold. It is noted that in the deposited complex structure, PCNA-127 is a Gly instead of the anticipated Lys. (B) Amino acid sequences of Srs2 PIP box and SIM in comparison to PIP and SIM from other yeast proteins. Consensus sequences in PIP and SIM are highlighted in green and purple, respectively. “h” represents amino acids with moderately hydrophobic side chains like L, I, V and M; “a” represents amino acids with highly hydrophobic, aromatic side chains like F and Y; and “x” represents any residues.
A Y2H assay (Fig. 6A) revealed that Pol30-2A reduced interaction with both Srs2 and Rad30; Pol30-4A affected Rad30 interaction but not Srs2 interaction, while Pol30-6A completely abolished interaction with both proteins.
FIG 6.
Phenotypes of Pol30 IDCL and C-terminal domain mutations. (A) Physical interaction between Pol30 mutations and Srs2 by a Y2H assay. Rad30 served as a reference. SRS2, SRS2-CT and RAD30 were cloned into pGBT9 (Gal4BD) and Pol30 mutations that disrupt either IDCL (L126A, I128A = 2A) or the C terminus (252-255AAAA = 4A), or both (=6A) were cloned into pGAD424 (Gal4AD). Experimental conditions were as described in Fig. 3. (B) The viability assay for pol30 mutants after plasmid shuffling. WXY939 cells transformed with YCpL-Pol30 derived plasmids were grown on YPD overnight to allow plasmid loss. Cells were then subjected to a serial dilution by spotting on SD-Leu to select those carrying YCpL-Pol30 based plasmid, on SD-Ura to select those carrying pBL211 (YCpU-POL30), and on 5-FOA to select cells that had lost the pBL211 plasmid. (C) Phenotypes of pol30-2A in rescuing rad5Δ or rad18Δ mutants from killing by DNA-damaging agents in a serial dilution assay. Experimental conditions were as described in Fig. 2. All plates were incubated at 30°C for 2 days before photography.
To assess effects of the above mutations on the rescue of rad5Δ and rad18Δ severe DNA damage sensitivity, we attempted to shuffle plasmids with these pol30 mutations. To our surprise, only pol30-2A cells were viable, albeit with reduced fitness, whereas pol30-4A and pol30-6A could not replace the plasmid carrying wild-type POL30 with a URA3 selectable marker (Fig. 6B). The above observations indicate that the PCNA C-terminal region plays an essential role, while its IDCL motif is important but dispensable for the cell viability.
The pol30-2A mutation alone caused moderate DNA-damage sensitivity and cannot rescue rad5Δ and rad18Δ cells from killing by DNA-damaging agents (Fig. 6C). Hence, a Pol30 mutation affecting interaction with both Srs2 and Rad30 cannot mimic the pol30-A171D phenotype.
pol30-I128A can rescue rad5Δ and rad18Δ in a fashion reminiscent of pol30-A171D.
To search for a Pol30-A171D mimetic mutation and validate our hypothesis, a group of less destructive mutations were designed either within or around Pol30 IDCL and C-terminal regions based on the SUMO-PCNA-Srs2-CT complex structure (Fig. 5A). As Pol30-I128 buries Srs2-L1156, Pol30-I128A is expected to weaken IDCL binding to Srs2. Pol30-F254A should affect Srs2 interaction with the PCNA C terminus, as the Pol30-F254 sidechain buries Srs2-M1150, while equivalent residues in other PIP-containing proteins are hydrophilic (Fig. 5B). Therefore, Pol30-F254A would leave the pocket accessible to solvent (Fig. 5A). Srs2-F1153 is an aromatic residue with a Pi system unique among PIP-containing proteins, and Pol30-R44 may stabilize Srs2-F1153 with a cation/Pi interaction because of their large aromatic hydrophobic sidechain (32). In addition, Pol30-R44 is in a recently defined central loop (residues 41 to 44) (33). Hence, Pol30-R44A should disrupt this interface outside PCNA IDCL and C terminus (Fig. 5A).
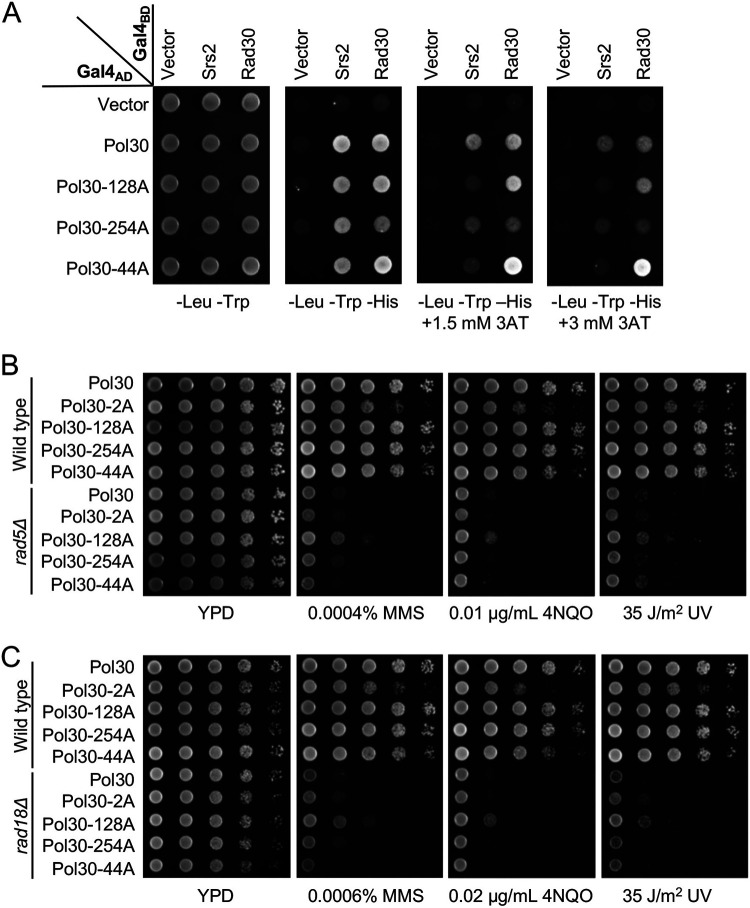
Like Pol30-A171D, Pol30-I128A reduced Srs2 but not Rad30 interaction in Y2H (Fig. 7A) and affinity pulldown (Fig. S3) assays. Meanwhile, Pol30-F254A reduced interaction with both Srs2 and Rad30, and Pol30-R44A decreased Srs2 interaction and enhanced Rad30 interaction (Fig. 7A). The above mutations were used to replace POL30 in rad5Δ and rad18Δ cells, and the DNA damage sensitivity assay showed that pol30-I128A rescued both rad5Δ (Fig. 7B) and rad18Δ (Fig. 7C) cells by about 10-fold, which was comparable to pol30-K164R but less effective than pol30-A171D.
FIG 7.
Phenotypes of selected pol30 amino acid substitution mutants. (A) Physical interaction between pol30 mutations and Srs2 or Rad30 by a Y2H assay. Experimental conditions were as described in Fig. 3. (B,C) Phenotypes of pol30 mutants in rescuing rad5Δ (B) and rad18Δ (C) mutants from killing by DNA-damaging agents in a serial dilution assay. Experimental conditions were as described in Fig. 2. All plates were incubated at 30°C for 2 days before photography.
In vitro interaction between Srs2 C terminus (Srs2-CT), Rad30-PIP and Pol30 or its mutant derivatives by a GST pulldown assay. Download FIG S3, DOCX file, 0.2 MB (166.8KB, docx) .
Copyright © 2023 Fan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
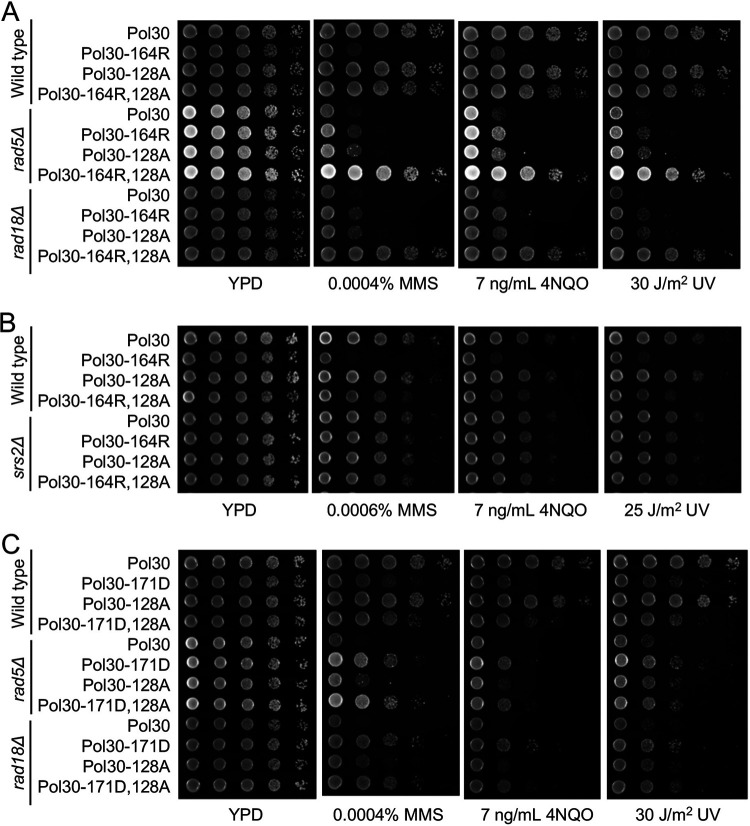
To further address the genetic relationship between pol30-I128A and pol30-K164R, the corresponding double mutation was created, and the two single mutations were found to be strongly synergistic. While each single mutation rescued rad5Δ or rad18Δ DNA-damage sensitivity by about 10-fold, the double mutation rescued rad5Δ and rad18Δ by at least 1,000-fold under all experimental conditions, to the level indistinguishable from the double mutant alone (Fig. 8A), reminiscent of the pol30-A171D,K164R mutation. As expected, the srs2 mutation is epistatic to pol30-I128A (Fig. 8B). Furthermore, the pol30-A171D,I128A double mutant behaved like the pol30-A171D and pol30-I128A single mutant (Fig. 8C), indicating that they rescued rad5 and rad18 severe sensitivity by the same mechanism. Like pol30-A171D, pol30-I128A also does not affect the PCNA-K164 sumoylation (Fig. S1, lanes 3 and 4). In summary, pol30-I128A mutant phenotypes are similar to pol30-A171D, allowing us to conclude that both pol30-I128A and pol30-A171D achieved rad5Δ and rad18Δ rescuing effects by specifically reducing interaction between PCNA and Srs2.
FIG 8.
Genetic interactions between pol30-I128A and relevant mutations by a serial dilution assay. (A) Genetic interaction between pol30-I128A and pol30-K164R in the rescue of rad5Δ and rad18Δ mutants. (B) Genetic interaction between pol30-I128A and pol30-K164R in the srs2 background. (C) Genetic interaction between pol30-I128A and pol30-A171D in the rescue of rad5Δ and rad18Δ mutants. Experimental conditions were as described in Fig. 2. All plates were incubated at 30°C for 2 days before photography.
Western blot analysis to detect PCNA post-translational modifications. Download FIG S1, DOCX file, 0.08 MB (81.5KB, docx) .
Copyright © 2023 Fan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Previous reports have established that sumoylated PCNA facilitates the Srs2 recruitment, leading to the inhibition of unwanted HR (8, 9). This is achieved through coordinated interaction of Srs2-PIP and Srs2-SIM with PCNA and SUMO, respectively, forming a tandem receptor (23). Inactivation of SRS2 can rescue the severe DNA damage sensitivity caused by malfunctional DDT in an HR dependent manner (18), a mechanism known as salvage HR (19, 20). Here, we report the isolation and systematic characterization of pol30-A171D that specifically reduces affinity for Srs2 to trigger salvage HR. First, an isolated pol30-A171D mutation can rescue not only rad5Δ, but also rad18Δ mutants from killing by DNA-damaging agents, indicating that this mutation induces salvage HR. Second, pol30-A171D does not affect PCNA sumoylationm and is synergistic with pol30-K164R in the rescuing, indicating that they act via distinct mechanisms. Third, since the srs2 mutation is epistatic to pol30-A171D and pol30-K164R, both Pol30 substitutions are speculated to reduce Srs2 recruitment. Finally, pol30-A171D reduces physical interaction with Srs2; pol30-K164R is known to abolish PCNA sumoylation, while the double mutation losses both means to bind Srs2-CT (Fig. 9A), which explains why the two mutations confer synergistic effects in preventing Srs2 recruitment to the replication fork. Furthermore, this study rules out a possibility that pol30-A171D affects the Pol30-K127 sumoylation, leading to the synergistic interaction with pol30-K164R. First, pol30-A171D rescues the severe DNA-damage sensitivity of rad5 and rad18, while pol30-K127R does not. Second, pol30-A171D and pol30-K127R are additive, indicating that they act in different pathways. Finally, Pol30-A171D reduces affinity for Srs2, while Pol30-K127R does not. The above observations support a notion that Pol30-K127 sumoylation can backup Pol30-K164 sumoylation in the recruitment of Srs2 (8, 9, 23) and hence both functions in the PCNA sumoylation branch (Fig. 9A).
FIG 9.
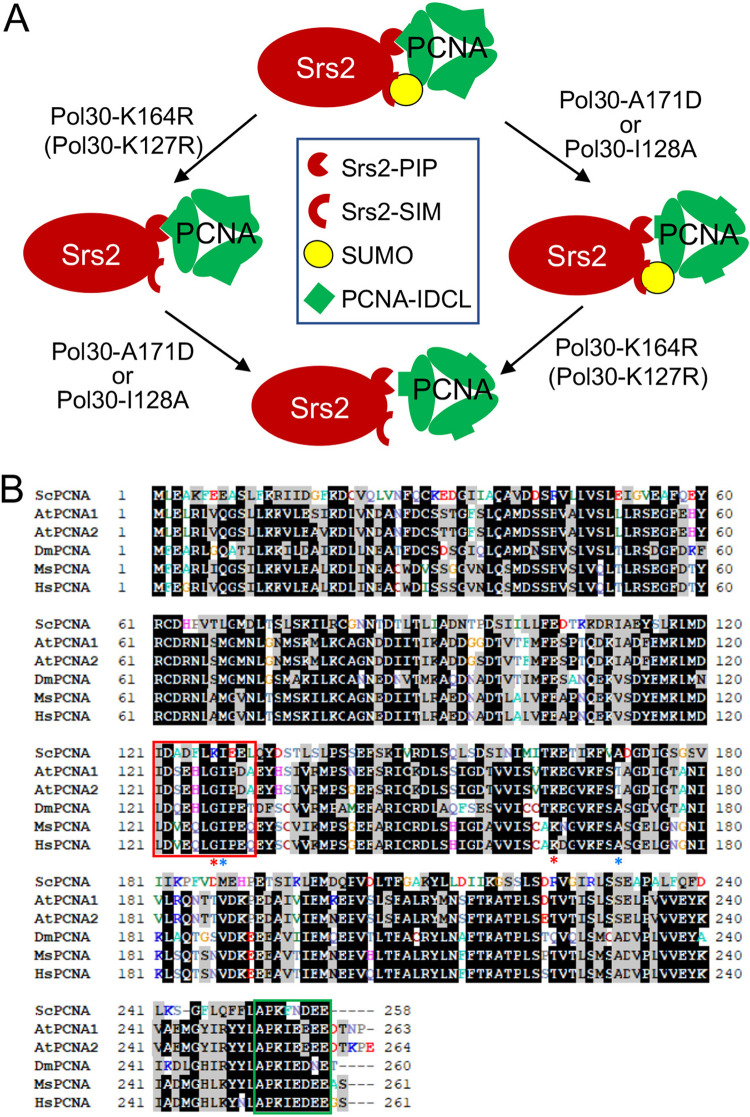
Critical PCNA residues in the recruitment of Srs2 and their conservation in other eukaryotes. (A) A working model on how Pol30-A171D/I128A and Pol30-K164R/K127R cooperatively disrupt physical interaction with Srs2. Srs2-CT contains a tandem receptor consisting of an atypical PIP box and a SIM domain that interact with PCNA IDCL and K164/K127-SUMO, respectively. While each single Pol30 mutation disrupt one such interaction, the corresponding double mutation fails to recruit Srs2, resulting in an equivalent srs2 mutant phenotype in the rescuing of severe rad5Δ/rad18Δ DNA-damage sensitivity. (B) Multiple sequence alignment among PCNAs from selected eukaryotic model organisms and human. Sc, Saccharomyces cerevisiae; At, Arabidopsis thaliana; Dm, Drosophila melanogaster; Mm, Mus musculus; and Hs, Homo sapiens. Residues highlighted by black are identical sequences and those by gray are conserved sequences. PCNA IDCL and C-terminal regions are in red and green boxes, respectively. Pol30-A171 and -I128 residues are indicated by blue asterisks and Pol30-K164 and -K127 residues are indicated by red asterisks.
One caveat for the above working model is that, based on the published SUMO-PCNA-Srs2-CT structure (23), Pol30-A171 is not located in the PCNA and Srs2-PIP box interface. To directly test our core hypothesis, a series of rationally designed pol30 mutant derivatives were created and their ability to induce salvage HR as well as genetic interactions with srs2 and pol30-K164R were examined, among which pol30-I128A functionally behaved like pol30-A171D. From the above complex structure, Pol30-I128 is located within the IDCL region in the PCNA-PIP interface, and the Pol30-I128A substitution specifically reduces Srs2 interaction but not Rad30 interaction, providing strong support to our working model (Fig. 9A). One can also see from the above structure that the sidechain of Pol30-A171 is buried in a pocket lined by four sidechains from Pol30-I158, -L151, -L154, -S155, and a β-sheet (Fig. 5A). Based on our collective observations and a speculation that Pol30-A171D also loosens PIP interaction, we predict that amino acid substitution from Pol30-A171 to a larger and hydrophilic Asp residue would destabilize the structure, which can be propagated to the Srs2-binding region(s).
Our Pol30 mutagenesis study reveals that PCNA binds Srs2-PIP and Rad30-PIP differently. First, Pol30-A171D selectively affects Srs2 but not Rad30 binding. Second, disruption of PCNA-IDCL by Pol30-2A reduces its interaction with both Srs2 and Rad30, while disruption of the PCNA C-terminal motif by Pol30-4A selectively interferes with Rad30 but not Srs2 interaction, indicating that the two PCNA regions preferentially bind different consensus sequences in the PIP box. Third, A single amino acid substitution in IDCL by Pol30-I128A has similar effects like Pol30-A171D, while Pol30-F254A in the C terminus only slightly reduces Rad30 binding. Finally, the Pol30-R44A substitution slightly reduces Srs2 interaction and enhances Rad30 binding, reminiscent of Pol30-K164R. The underlying mechanism appears to be that most PIP-boxes adopt a β-stranded structure at the N terminus followed by a 310 helix at the C terminus, in which Cdc9-F45 is proximal to Pol30-L126 and I128 (27), whereas Srs2 residues 1153FSQL1156 adopt two turns of α-helix instead, in which Srs2-L1156 is proximal to Pol30-I128 and L131 (23). Our observation that Pol30-I128A reduces Srs2 interaction is consistent with a report that Srs2-L1156A also reduces its affinity for Pol30 (23). Apparently, Pol30-I128A would have only a moderate if any effect on the interaction with “FF” motif-containing PIP box proteins. In summary, if a Pol30 mutation only affects Srs2 interaction but not classical PIP box proteins (e.g., A171D and I128A), it could compromise Srs2 recruitment to the replication fork and activate the salvage HR. If a mutation affects both Rad30 and Srs2 (e.g., F254A) or only Rad30 but not Srs2 interaction (e.g., 2A), it cannot rescue the severe DNA-damage sensitivity of DDT mutants. Apparently, interaction with PIP-box proteins is an essential function for PCNA, as disruption of its C-terminal domain cannot maintain cell viability, a conclusion consistent with the structural and biochemical analyses (27). Fortunately, the Srs2-PIP is perhaps unique and hence targeted mutations in PCNA can be made to specifically disrupt its interaction without severe consequences on other PIP-box proteins.
Why have cells evolved such partial PIP boxes to be involved in the interaction with PCNA? One can imagine that proteins containing a full PIP box, including the Cdc9 DNA ligase, Polδ, and the Polε catalytic subunit Pol2, have relatively high affinity for, and constantly travel with, PCNA during replication. On the other hand, a partial PIP box may facilitate regulated association and dissociation from PCNA. For example, Srs2 is efficiently recruited by PCNA only when it is sumoylated (8, 9). Polη (Rad30) is recruited to the stalled replication fork when PCNA is monoubiquitinated at the same K164 residue (3) and can be dissociated when PCNA is no longer ubiquitinated (34). Interestingly, in addition to its genome replication role, Polδ is also involved in DDT (35 to 37), and some of its subunits contain partial PIP boxes (26). In response to DNA damage that blocks replication, a polymerase switch between Polδ (Pol3, Pol31 and Pol32) and Polζ4 (Rev3, Rev7, Pol31 and Pol32) takes place (38, 39). Whether this process involves PIP box regulation remains elusive.
Amino acid sequence alignment (Fig. 9B) reveals that PCNA IDCL and C-terminal regions are highly conserved throughout eukaryote organisms, from yeasts to human. The conserved residues include both Pol30-A171 and -I128 (Fig. 9B), suggesting that the regulatory mechanism as described in this study also applies to other eukaryotic organisms, although true Srs2 orthologs have not yet been identified in higher eukaryotes. Hence, this study sheds light on the investigation of DDT pathway regulation in higher eukaryotes, including plants and mammals.
MATERIALS AND METHODS
Yeast strains and cell culture.
S. cerevisiae cells were grown either in rich yeast-extract peptone dextrose (YPD) medium (1% yeast extract, 2% peptone, 2% glucose) or a synthetic dextrose (SD) medium (0.17% yeast nitrogen base without amino acids, 5% ammonium sulfate, 2% glucose supplemented with appropriate amino acids and bases). 2% agar was added when making plates. Plates containing 5-fluoroorotic acid (5-FOA) were made as previously described (29) to select for the ura3 auxotroph.
Yeast strains used in this study are listed in Table S1. All strains used in this study were isogenic to HK578-10D except PJ69-4a (40). Gene deletion mutants were created by a one-step gene deletion method (41) using disruption cassettes as described (42), and the target gene deletion was confirmed by genomic PCR, followed by phenotypic analysis. Plasmids were transformed into yeast cells following a LiAc/single-strand carrier DNA/PEG method (43).
Saccharomyces strains used in this study. Download Table S1, DOCX file, 0.02 MB (19.4KB, docx) .
Copyright © 2023 Fan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmid construction and site-specific mutagenesis.
Plasmid YCpL-Pol30 was constructed by cloning the POL30 open reading frame (ORF) along with its own promoter and terminator sequences into YCplac111 (YCp, LEU2) (44) as previously described (45). Site-specific pol30 mutants were created in plasmid YCpL-Pol30 by a modified Quick Change method (46) using primers as shown in Table S2. The resulting plasmids were confirmed by sequencing the entire insert.
Oligonucleotides used in this study. Download Table S2, DOCX file, 0.02 MB (22.4KB, docx) .
Copyright © 2023 Fan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
For the yeast two-hybrid (Y2H) assay, indicated yeast genes or fragments were amplified by PCR using primers containing restriction enzyme cleavage sites as listed in Table S2, cleaved by the corresponding restriction enzymes and cloned into either pGBT9 or pGAD424 (47) as Gal4BD or Gal4AD fusion, respectively. The Escherichia coli transformants were screened by colony PCR and the resulting plasmids were further confirmed by sequencing the entire insert.
To produce recombinant His6-tagged Pol30, plasmid pET-Pol30 was constructed by cloning the POL30 ORF into BamHI-SacI sites of pET30a. To produce recombinant GST-tagged Srs2 C-terminal (Srs2-CT) domain, the DNA sequence encoding Srs2 residues 1107 to 1174 was amplified by primers Srs2-CT-BamHI-F and Srs2-CT-EcoRI-R (Table S2) and cloned into the BamHI-EcoRI sites of pGEX6. To produce recombinant GST-tagged Rad30-PIP, DNA sequences encoding Rad30 residues 515 to 632 was amplified by primers Rad30-BamHI-F and Rad30-EcoRI-R (Table S2) and cloned into the BamHI-EcoRI sites of pGEX6. All inserts in the cloned plasmids were confirmed by sequencing.
Plasmid shuffling.
A plasmid shuffling method (29) was used to replace plasmid-borne wild-type POL30 with pol30 mutants. Briefly, yeast strain WXY939 and its isogenic mutants contain a chromosomal pol30Δ::HIS3 allele and the cell viability was maintained by a plasmid pBL211 (YCp, URA3, POL30) (48), a gift from P. Burgers (University of Washington, St. Louis). To shuffle plasmids, WXY939 or its mutant cells were transformed with YCpL-Pol30 derived plasmids, resulting in transformants carrying two plasmids. The transformed cells were cultured in liquid YPD overnight, washed twice with sterile H2O, properly diluted and then 0.1 mL culture was spread on a 5-FOA plate. The plate was incubated at 30°C for 3 days and individual colonies were picked and streaked onto a fresh 5-FOA plate. Since the URA3 gene product converts 5-FOA into a compound toxic to yeast cells (49), cells are able to grow only after they have lost the URA3-containing plasmid. Hence, the 5-FOA resistant cells were expected to only carry the YCpL-Pol30 plasmid or its mutant forms, whose phenotypes were then assessed. If the transformed cells do not form colonies on the 5-FOA plate, it is an indication that the pol30 mutation carried by the YCpL-Pol30 plasmid cannot support the POL30 essential function.
Yeast cell survival assays.
The gradient plate and serial dilution assays were performed as previously described (50) to assess relative sensitivity of yeast cells to DNA-damaging agents. Briefly, for the gradient plate assay, overnight cultures were printed using a microscopic slide onto a set of two-layer YPD agar plates containing MMS concentration gradients. The plates were incubated for 2 to 3 days at 30°C before photography. For the serial dilution assay, overnight yeast cultures were used to make a set of 10-fold dilutions and then 4.5 μL samples were spotted on freshly made YPD agar plates containing different concentrations of MMS or 4NQO. UV treatment was achieved by exposing yeast cells in the spotted plate to UV irradiation in a UV cross-linker (Stratagene SS-UV1800) at given doses. All plates were incubated for 2 to 3 days at 30°C in the dark before photography. For both assays, only representative and informative plates were presented.
Yeast two-hybrid assay.
Plasmids expressing Gal4AD and Gal4BD fusion proteins to be tested were cotransformed into yeast strain PJ69-4a. Transformants were allowed to grow at 30°C on an SD-Leu-Trp plate for 2 to 3 days, after which at least two groups of individual colonies were cultured and then spotted on an SD-Leu-Trp plate as control and on SD-Leu-Trp-His plates with or without certain concentrations of a histidine biosynthesis inhibitor 1, 2, 4-amino triazole (3AT) (51) to interrogate physical interaction between the two expressed gene products based on the relative growth on the selective plates. All plates were incubated for 2 to 6 days at 30°C before photography. Only representative and informative plates were shown.
GST pulldown assay.
E. coli BL21 cells expressing His6-tagged Pol30 or GST-tagged Srs2-CT Rad30-PIP were induced by 0.1 mM IPTG at 16°C for 16 to 18 h. Cells were harvested by centrifugation, resuspended in corresponding His6 lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 30 mM imidazole) or GST lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 10 mM β-mercaptoethanol) and then homogenized in a cell disruptor (Constant Cell Disruption Systems, CF1) at 25 lb/in2 by two passes. Cells lysates were centrifuged at 16,000 g for 60 min. His6-Pol30 was affinity purified by Ni Sepharose (Cytiva, 17531801) and then eluted from the Ni Sepharose by 5-fold bed volume elution buffer (20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 500 mM imidazole, 20% glycerol). GST-Srs2-CT and GST-Rad30-PIP were affinity purified by glutathione Sepharose (Cytiva, GE17-0756-01) in a GST stock buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 10 mM β-mercaptoethanol, 20% glycerol). The above proteins were either freshly used or frozen in liquid nitrogen and stored at −80°C. Equal molars of GST-Srs2/Rad30 and His6-Pol30 were coincubated overnight at 4°C with gentle shaking. Anti-GST (Sigma, G7781-25UL, 1:1000), anti-His6 (New England Biolabs, 12698, 1:1000) primary antibodies and the anti-Rabbit (Bio-Rad, 1706515, 1: 3000) secondary antibody were used in immunoblotting.
Western blot analyses.
Overnight cultured yeast cells were used to inoculate 50 mL fresh YPD at 1:20 dilution and the incubation continued at 30°C until OD600nm =0.35 to 0.4. Cells were pelleted by centrifugation, washed with ddH2O and resuspended in 0.5 mL ddH2O, to which equal volume 0.2 M NaOH was added and incubated at 24°C for 15 min before adding 150 μL buffer containing 60 mM Tris-HCl, pH 6.8, 4% SDS, 0.01% bromophenol blue, 5% glycerol and 4% β-mercaptoethanol (52). The sample was boiled for 10 min, centrifuged at 15,000 rpm for 2 min and the supernatant was subjected to SDS-PAGE and Western blotting by using anti-PCNA (ab70472, Abcam, 1:1000) and anti-Pgk1 (a gift from Wei Li, Institute of Microbiology, Chinese Academic of Sciences) primary antibodies, and anti-mouse (Thermo Scientific 3140, 1:4000) and anti-rabbit (Bio-Rad, 1706515, 1: 5000) secondary antibodies, respectively.
Protein structural and bioinformatics analyses.
The SUMO-PCNA-Srs2-CT complex structure (23) was obtained from the Protein Data Bank (PDB, https://www.rcsb.org/) entry 3v62 and displayed by using Molscript (53). The Cdc9-PCNA complex structure (27) was obtained from the PDB entry 2od8.
PCNA sequences were downloaded from NCBI (https://www.ncbi.nlm.nih.gov). Multiple alignments were performed and presented by BioEdit 7.2 downloaded from https://bioedit.software.informer.com/7.2/.
ACKNOWLEDGMENTS
We thank P. Burgers for plasmid pBL211, P. James for yeast strain PJ69-4a and W. Li for the anti-Pgk1 antibody.
This work was supported by Natural Sciences and Engineering Research Council of Canada Discovery Grants RGPIN-2019-05604, College of Medicine CoMRAD, and the National Natural Science Foundation of China operating grant 31670068 to W.X.
We have no conflicts of interest to declare.
Contributor Information
Wei Xiao, Email: wei.xiao@usask.ca.
Fred M. Winston, Harvard Medical School
REFERENCES
- 1.Barbour L, Xiao W. 2003. Regulation of alternative replication bypass pathways at stalled replication forks and its effects on genome stability: a yeast model. Mutat Res 532:137–155. doi: 10.1016/j.mrfmmm.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 2.Pastushok L, Xiao W. 2004. DNA postreplication repair modulated by ubiquitination and sumoylation. Adv Protein Chem 69:279–306. doi: 10.1016/S0065-3233(04)69010-3. [DOI] [PubMed] [Google Scholar]
- 3.Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, Hofmann K, Dikic I. 2005. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 4.Guo C, Sonoda E, Tang TS, Parker JL, Bielen AB, Takeda S, Ulrich HD, Friedberg EC. 2006. REV1 protein interacts with PCNA: significance of the REV1 BRCT domain in vitro and in vivo. Mol Cell 23:265–271. doi: 10.1016/j.molcel.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 5.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 6.Ball LG, Zhang K, Cobb JA, Boone C, Xiao W. 2009. The yeast Shu complex couples error-free post-replication repair to homologous recombination. Mol Microbiol 73:89–102. doi: 10.1111/j.1365-2958.2009.06748.x. [DOI] [PubMed] [Google Scholar]
- 7.Mankouri HW, Ngo HP, Hickson ID. 2007. Shu proteins promote the formation of homologous recombination intermediates that are processed by Sgs1-Rmi1-Top3. Mol Biol Cell 18:4062–4073. doi: 10.1091/mbc.e07-05-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. 2005. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell 19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. 2005. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 10.Lawrence CW, Christensen RB. 1979. Metabolic suppressors of trimethoprim and ultraviolet light sensitivities of Saccharomyces cerevisiae rad6 mutants. J Bacteriol 139:866–876. doi: 10.1128/jb.139.3.866-876.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aboussekhra A, Chanet R, Zgaga Z, Cassier-Chauvat C, Heude M, Fabre F. 1989. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res 17:7211–7219. doi: 10.1093/nar/17.18.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rong L, Palladino F, Aguilera A, Klein HL. 1991. The hyper-gene conversion hpr5-1 mutation of Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics 127:75–85. doi: 10.1093/genetics/127.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 14.Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 15.Broomfield S, Xiao W. 2002. Suppression of genetic defects within the RAD6 pathway by srs2 is specific for error-free post-replication repair but not for damage-induced mutagenesis. Nucleic Acids Res 30:732–739. doi: 10.1093/nar/30.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedl AA, Liefshitz B, Steinlauf R, Kupiec M. 2001. Deletion of the SRS2 gene suppresses elevated recombination and DNA damage sensitivity in rad5 and rad18 mutants of Saccharomyces cerevisiae. Mutat Res 486:137–146. doi: 10.1016/s0921-8777(01)00086-6. [DOI] [PubMed] [Google Scholar]
- 17.Ulrich HD. 2001. The srs2 suppressor of UV sensitivity acts specifically on the RAD5- and MMS2-dependent branch of the RAD6 pathway. Nucleic Acids Res 29:3487–3494. doi: 10.1093/nar/29.17.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schiestl RH, Prakash S, Prakash L. 1990. The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics 124:817–831. doi: 10.1093/genetics/124.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Branzei D, Szakal B. 2016. DNA damage tolerance by recombination: molecular pathways and DNA structures. DNA Repair (Amst) 44:68–75. doi: 10.1016/j.dnarep.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan L, Bi T, Wang L, Xiao W. 2020. DNA-damage tolerance through PCNA ubiquitination and sumoylation. Biochem J 477:2655–2677. doi: 10.1042/BCJ20190579. [DOI] [PubMed] [Google Scholar]
- 21.Moldovan GL, Pfander B, Jentsch S. 2007. PCNA, the maestro of the replication fork. Cell 129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Warbrick E, Heatherington W, Lane DP, Glover DM. 1998. PCNA binding proteins in Drosophila melanogaster: the analysis of a conserved PCNA binding domain. Nucleic Acids Res 26:3925–3932. doi: 10.1093/nar/26.17.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Armstrong AA, Mohideen F, Lima CD. 2012. Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature 483:59–63. doi: 10.1038/nature10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boehm EM, Powers KT, Kondratick CM, Spies M, Houtman JC, Washington MT. 2016. The proliferating cell nuclear antigen (PCNA)-interacting protein (PIP) motif of DNA polymerase eta mediates its interaction with the C-terminal domain of Rev1. J Biol Chem 291:8735–8744. doi: 10.1074/jbc.M115.697938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L. 2001. Interaction with PCNA is essential for yeast DNA polymerase eta function. Mol Cell 8:407–415. doi: 10.1016/S1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 26.Acharya N, Klassen R, Johnson RE, Prakash L, Prakash S. 2011. PCNA binding domains in all three subunits of yeast DNA polymerase delta modulate its function in DNA replication. Proc Natl Acad Sci USA 108:17927–17932. doi: 10.1073/pnas.1109981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vijayakumar S, Chapados BR, Schmidt KH, Kolodner RD, Tainer JA, Tomkinson AE. 2007. The C-terminal domain of yeast PCNA is required for physical and functional interactions with Cdc9 DNA ligase. Nucleic Acids Res 35:1624–1637. doi: 10.1093/nar/gkm006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu X, Lin A, Zhou C, Blackwell SR, Zhang Y, Wang Z, Feng Q, Guan R, Hanna MD, Chen Z, Xiao W. 2016. Involvement of budding yeast Rad5 in translesion DNA synthesis through physical interaction with Rev1. Nucleic Acids Res 44:5231–5245. doi: 10.1093/nar/gkw183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan L, Xiao W. 2021. Study essential gene functions by plasmid shuffling, p 53–62. In Xiao W (ed), Yeast Protocols, 4th Ed, vol 2196. Springer Nature Press, New York, USA. [DOI] [PubMed] [Google Scholar]
- 30.Moldovan GL, Pfander B, Jentsch S. 2006. PCNA controls establishment of sister chromatid cohesion during S phase. Mol Cell 23:723–732. doi: 10.1016/j.molcel.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 31.Branzei D, Seki M, Enomoto T. 2004. Rad18/Rad5/Mms2-mediated polyubiquitination of PCNA is implicated in replication completion during replication stress. Genes Cells 9:1031–1042. doi: 10.1111/j.1365-2443.2004.00787.x. [DOI] [PubMed] [Google Scholar]
- 32.McGaughey GB, Gagne M, Rappe AK. 1998. pi-Stacking interactions. J Biol Chem 273:15458–15463. doi: 10.1074/jbc.273.25.15458. [DOI] [PubMed] [Google Scholar]
- 33.Bhardwaj VK, Purohit R. 2020. A new insight into protein-protein interactions and the effect of conformational alterations in PCNA. Int J Biol Macromol 148:999–1009. doi: 10.1016/j.ijbiomac.2020.01.212. [DOI] [PubMed] [Google Scholar]
- 34.Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D'Andrea AD. 2006. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol 8:341–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 35.Torres-Ramos CA, Prakash S, Prakash L. 1997. Requirement of yeast DNA polymerase delta in post-replicational repair of UV-damaged DNA. J Biol Chem 272:25445–25448. doi: 10.1074/jbc.272.41.25445. [DOI] [PubMed] [Google Scholar]
- 36.Huang ME, Rio AG, Galibert MD, Galibert F. 2002. Pol32, a subunit of Saccharomyces cerevisiae DNA polymerase delta, suppresses genomic deletions and is involved in the mutagenic bypass pathway. Genetics 160:1409–1422. doi: 10.1093/genetics/160.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanna M, Ball LG, Tong AH, Boone C, Xiao W. 2007. Pol32 is required for Pol zeta-dependent translesion synthesis and prevents double-strand breaks at the replication fork. Mutat Res 625:164–176. doi: 10.1016/j.mrfmmm.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 38.Baranovskiy AG, Lada AG, Siebler HM, Zhang Y, Pavlov YI, Tahirov TH. 2012. DNA polymerase delta and zeta switch by sharing accessory subunits of DNA polymerase delta. J Biol Chem 287:17281–17287. doi: 10.1074/jbc.M112.351122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Makarova AV, Stodola JL, Burgers PM. 2012. A four-subunit DNA polymerase zeta complex containing Pol delta accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res 40:11618–11626. doi: 10.1093/nar/gks948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.James P, Halladay J, Craig EA. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rothstein RJ. 1983. One-step gene disruption in yeast. Methods Enzymol 101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 42.Gonzalez C, Perdomo G, Tejera P, Brito N, Siverio JM. 1999. One-step, PCR-mediated, gene disruption in the yeast Hansenula polymorpha. Yeast 15:1323–1329. doi:. [DOI] [PubMed] [Google Scholar]
- 43.Gietz RD. 2014. Yeast transformation by the LiAc/SS carrier DNA/PEG method. Methods Mol Biol 1205:1–12. doi: 10.1007/978-1-4939-1363-3_1. [DOI] [PubMed] [Google Scholar]
- 44.Gietz RD, Sugino A. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 45.Pastushok L, Hanna M, Xiao W. 2010. Constitutive fusion of ubiquitin to PCNA provides DNA damage tolerance independent of translesion polymerase activities. Nucleic Acids Res 38:5047–5058. doi: 10.1093/nar/gkq239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng L, Baumann U, Reymond JL. 2004. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fields S, Song O. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 48.Ayyagari R, Impellizzeri KJ, Yoder BL, Gary SL, Burgers PM. 1995. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol Cell Biol 15:4420–4429. doi: 10.1128/MCB.15.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boeke JD, LaCroute F, Fink GR. 1984. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet 197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 50.Xu X, Lambrecht AD, Xiao W. 2014. Yeast survival and growth assays, p 183–191. In Xiao W (ed), Yeast Protocols, 3rd Ed, vol 1163. Humana Press, New York. [DOI] [PubMed] [Google Scholar]
- 51.Walhout AJ, Vidal M. 1999. A genetic strategy to eliminate self-activator baits prior to high-throughput yeast two-hybrid screens. Genome Res 9:1128–1134. doi: 10.1101/gr.9.11.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsuo Y, Asakawa K, Toda T, Katayama S. 2006. A rapid method for protein extraction from fission yeast. Biosci Biotechnol Biochem 70:1992–1994. doi: 10.1271/bbb.60087. [DOI] [PubMed] [Google Scholar]
- 53.Esnouf RM. 1997. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J Mol Graph Model 15:132–133. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genetic interactions between pol30-K164R and pol30-K127R in rescuing severe DNA-damage sensitivities of rad5 and rad18 mutants by a serial dilution assay. Download FIG S2, DOCX file, 0.2 MB (217.9KB, docx) .
Copyright © 2023 Fan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In vitro interaction between Srs2 C terminus (Srs2-CT), Rad30-PIP and Pol30 or its mutant derivatives by a GST pulldown assay. Download FIG S3, DOCX file, 0.2 MB (166.8KB, docx) .
Copyright © 2023 Fan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Western blot analysis to detect PCNA post-translational modifications. Download FIG S1, DOCX file, 0.08 MB (81.5KB, docx) .
Copyright © 2023 Fan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Saccharomyces strains used in this study. Download Table S1, DOCX file, 0.02 MB (19.4KB, docx) .
Copyright © 2023 Fan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Oligonucleotides used in this study. Download Table S2, DOCX file, 0.02 MB (22.4KB, docx) .
Copyright © 2023 Fan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.