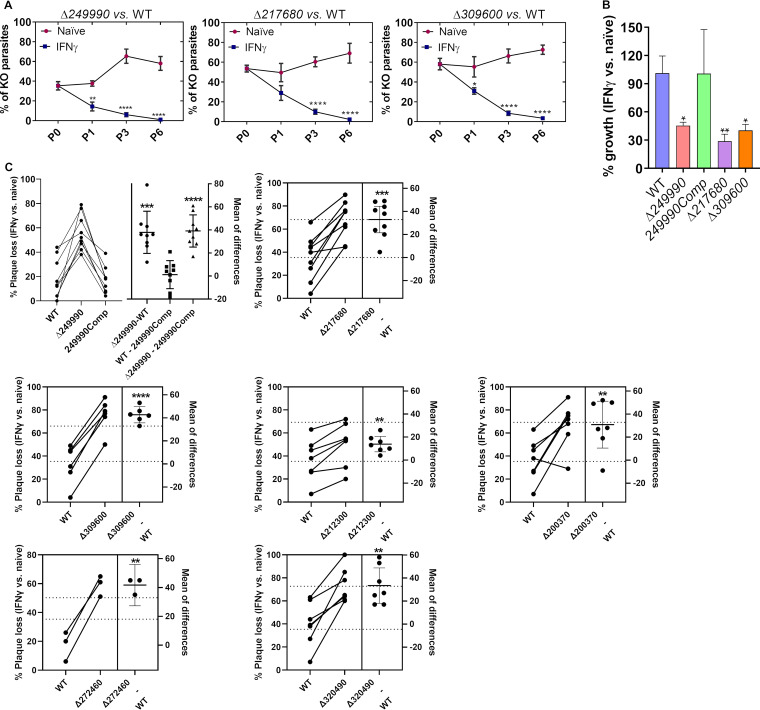
FIG 2.
Six of the top candidate genes identified in the CRISPR screen determine fitness in IFNγ-stimulated HFFs. (A) Indicated GFP-expressing knockout (KO) strains (numbers shown are TGGT1_ T. gondii gene ID numbers) generated in an RH (type I) luciferase-expressing background were mixed in a 1:1 ratio with luciferase-expressing wild-type (WT) parasites and passaged for 6 passages on naive or IFNγ-stimulated HFFs. The percentage of KO parasites was determined at passage 0, 1, 3, and 6 by plaque assays. Two-way ANOVA followed by Sidak’s multiple-comparison test was used for statistical analysis. (B) Naive or IFNγ-stimulated (20 U/mL) HFFs were infected with indicated parasite strains (249990Comp is Δ249990 complemented with a WT copy of 249990) and 24 hpi, parasite luciferase was quantified with a luminometer. Indicated is the percentage of luciferase in IFNγ-stimulated HFFs compared to unstimulated HFFs for each strain. Two-way ANOVA followed by Dunnett’s multiple-comparison test was used for statistical analysis. (C) Percentage decrease in number of lysis plaques formed in IFNγ-stimulated versus naive HFFs is plotted for indicated strains. On the right of each graph, the mean difference ±SD between KO and WT is indicated. Each data point is from a separate biological experiment (but some WT data points are shared among graphs from when multiple KO parasites were compared at the same time to WT). Paired Student's t test was used for the comparisons between WT and KO parasites, while ANOVA was used to compare WT, Δ249990, and 249990Comp. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 versus WT.