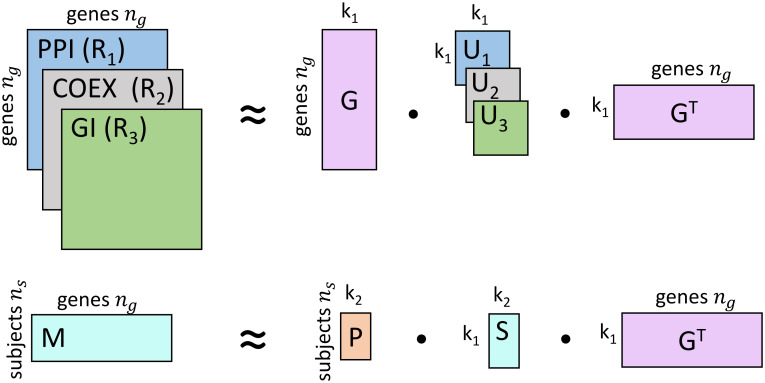
Fig 2. Our data-integration framework.
We used the NMTF method to integrate four different data types: germline mutations of the five subjects presented in , protein-protein interactions (PPIs) from BioGRID presented in , coexpressions (COEXs) from COXPRESdb presented in and genetic interactions (GIs) from [14] presented in . M is simultaneously decomposed into the product of three lower dimensional matrix factors, P, S and GT; molecular networks, Ri, are simultaneously decomposed into the product of three factors, G, Ui and GT, as detailed in Materials and methods. k1 and k2 are the gene and the patient clusters, respectively.