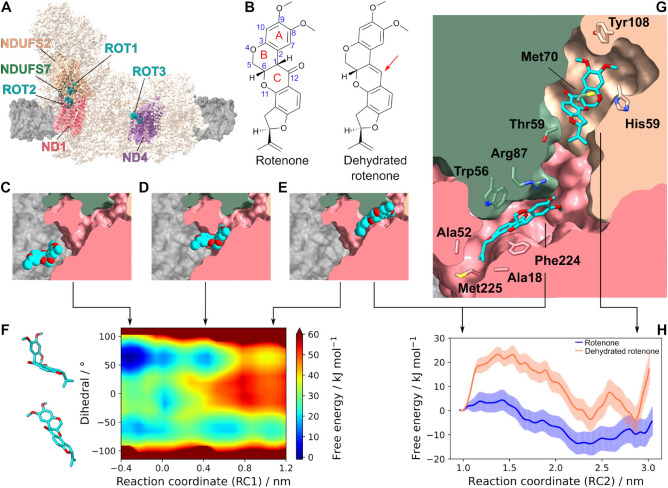
Figure 1.
Mechanism of rotenone binding into respiratory complex I. (A) Structure of complex I is shown with subunits ND1, ND4, NDUFS2 and NDUFS7 colored respectively in pink, purple, peach and green, with the membrane in gray. Three rotenone binding modes (ROT1, ROT2 and ROT3) observed in cryoEM models8,9 are displayed in cyan spheres. (B) Chemical structures of rotenone (left) and dehydrated rotenone (right), with a red arrow indicating C1C12 double bonding. Rotenone is shown in cyan (carbon) and red (oxygen) spheres on the (C) membrane, (D) narrow entrance and (E) ROT2 binding mode along the entry process. (F) Two-dimensional free energy profile for the entry process (corresponding to panels C D E). Rotenone structures in bent (dihedral ) and straight () conformations are shown on the left. (G) The Q-channel, with rotenone in binding modes ROT2 and ROT1. Two molecules are displayed here for visualisation, but only one was present during simulations. (H) Free energy profile for ligand transit from channel central region (ROT2) to Q-redox site (ROT1) for rotenone (blue) and dehydrated rotenone (orange). Uncertainties are shown in colored shadows. Arrows from panels (C–E and G point to the corresponding reaction coordinate in panels (F) (RC1) and (H) (RC2).