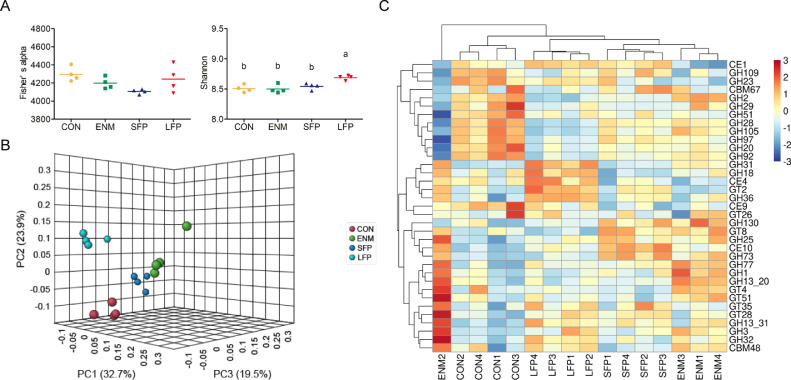
Figure 2.
Comparison of microbial carbohydrate-active enzyme gene structure in the cecal digesta. (A) Scatter plot of alpha diversity of microbial carbohydrate-active enzyme gene composition in the CON, ENM, SFP, and LFP groups. Each bar represents mean ± standard deviation (n = 4). Different superscripts indicate significant differences between groups. (B) PCoA of the microbial carbohydrate-active enzyme gene composition in the CON, ENM, SFP, and LFP groups (n = 4). (C) Heatmap of microbial carbohydrate-active enzyme gene abundance in the cecal digesta. Abundance distribution of 35 dominant microbial carbohydrate-active enzyme genes (Y-axis) across all samples (X-axis) is displayed (n = 4). The values are normalized using the Z-score. Abbreviations: CON, basal diet; ENM, basal diet supplemented with enramycin; LFP, Basal diet supplemented with B. licheniformis–fermented products; PCoA, principal coordinate analysis; SFP, Basal diet supplemented with B. subtilis–fermented products.