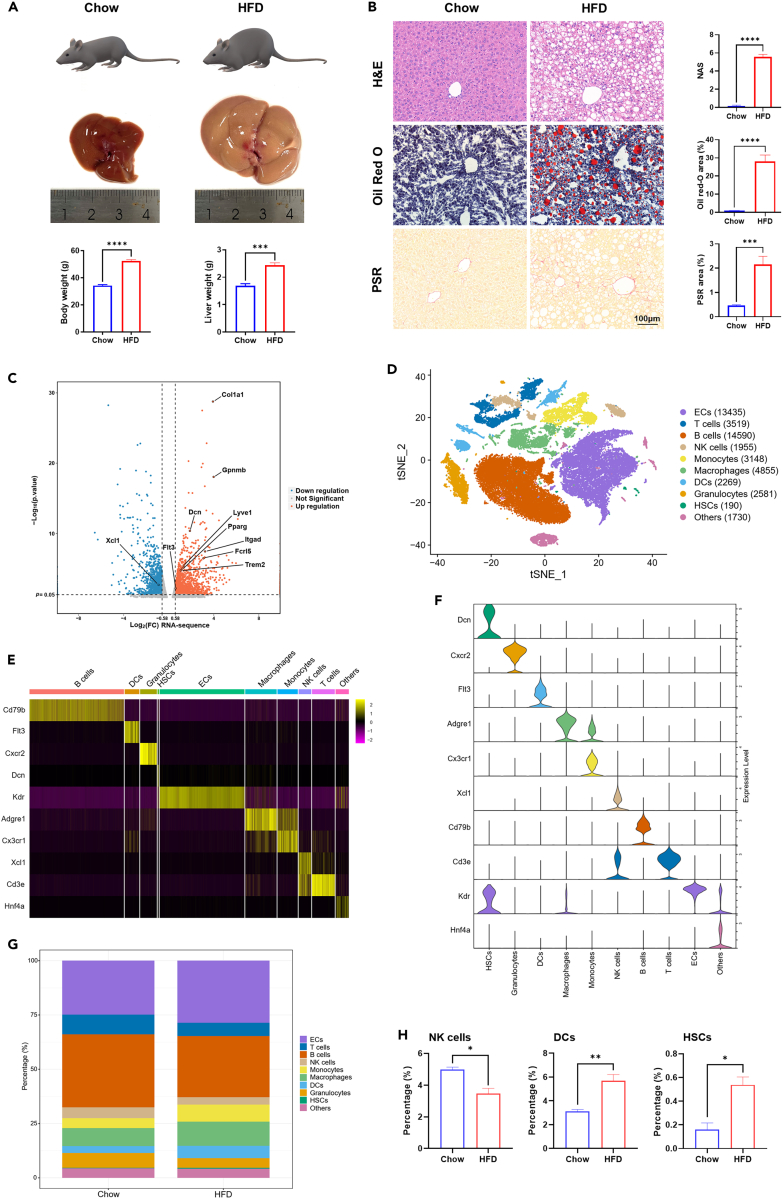
Figure 1.
Transcriptional analysis of liver NPCs on the bulk and single-cell level in healthy and NASH mice
(A) Body weight and liver weight of chow and HFD mice. Mice were fed with chow diet (Chow) and HFD diet supplemented with fructose and sucrose in the drinking water (HFD), respectively for 20 weeks.
(B) Hepatic steatosis, lipid accumulation, inflammation, and fibrosis in each group were evaluated by hematoxylin-eosin (H&E), oil red O, picrosirius red (PSR), and NAFLD activity score (NAS). Scale bar, 100 μm. The statistics were shown on the right.
(C) Differentially expressed genes between chow and HFD group on the bulk level. Genes up- or downregulated by > 1.5-fold were shown in orange and blue, respectively, while those ≤ 1.5-fold were shown in gray.
(D) t-SNE visualization of scRNA-seq results. NPCs were classified into 10 distinct types. Cell counts for each cell type were shown in parentheses.
(E) Heatmap of cell type specific marker genes.
(F) Representative marker gene expression in each cell type.
(G) Percentage of each cell type in NPCs isolated from livers of chow and HFD mice.
(H) Differences in the percentage of hepatic NK cells, DCs, and HSCs between chow and HFD group. n = 3–5 per group. The data are expressed as mean ± SEM. Statistical significance is shown as ∗ for p < 0.05, ∗∗ for p < 0.01, ∗∗∗ for p < 0.001, and ∗∗∗∗ for p < 0.0001, as evaluated by Student’s t test.