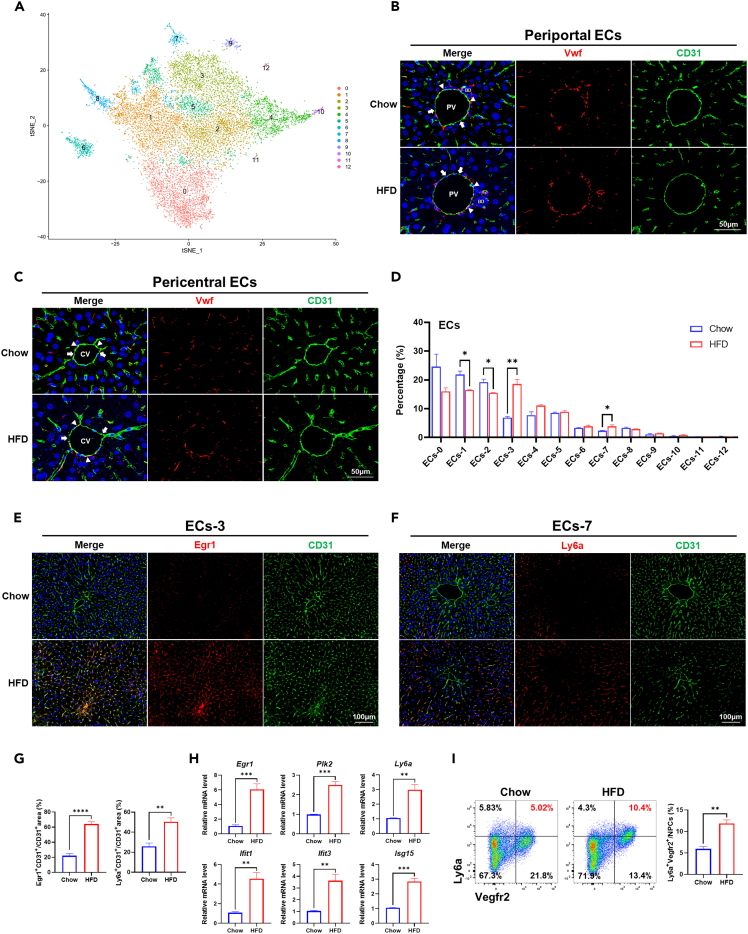
Figure 3.
Heterogeneity of ECs in mice liver
(A) t-SNE visualization of re-clustered hepatic ECs.
(B) Vwfhi and Vwflow ECs in the periportal area in chow and HFD mice liver. PV, portal vein. BD, bile duct. Arrowheads indicated Vwfhi ECs, and arrows indicated Vwflow ECs. Scale bar, 50 μm.
(C) Vwfhi and Vwflow ECs in the pericentral area in chow and HFD mice liver. CV: Central vein. Arrowheads indicate Vwfhi ECs, and arrows indicated Vwflow ECs. Scale bar, 50 μm.
(D) The percentage of each cluster in hepatic ECs in chow and HFD group.
(E) Detection of Egr1+ ECs (ECs-3) in chow and HFD mice liver by immunofluorescence staining. Scale bar, 100 μm.
(F) Detection of Ly6a+ ECs (ECs-7) in chow and HFD mice liver by immunofluorescence staining. Scale bar, 100 μm.
(G) The statistics of CD31+Egr1+ double-positive and CD31+Ly6a+ double-positive areas in chow and HFD mice liver.
(H) Expression of marker genes for ECs-3 (Egr1, Plk2) and ECs-7 (Ly6a, Ifit1, Ifit3, Isg15) in liver NPCs of chow and HFD group at mRNA level.
(I) Detection of Ly6ahi ECs (ECs-7) in chow and HFD mice liver by flow cytometry. Ly6ahi Vegfr2hi ECs were located on the top right panel of each plot, and their percentages in liver NPCs were shown in red. The statistics were shown on the right. n = 3–5 per group. The data are expressed as mean ± SEM. Statistical significance is shown as ∗ for p < 0.05, ∗∗ for p < 0.01, ∗∗∗ for p < 0.001, and ∗∗∗∗ for p < 0.0001, as evaluated by Student’s t test.