Abstract
Metabolite identification represents a major challenge, and opportunity, for biochemistry – the challenge of determining the structures of unknown metabolites and the opportunity of expanding our biochemical knowledge base. The collective characterization and quantification of pools of metabolites in living organisms, with its many successes, represents the foundation of metabolism’s rebirth in the 21st century. Historically, an enduring obstacle in the metabolic sciences has been characterizing newly observed metabolites; metabolites that have a distinct elemental composition (combustion analysis) or a mass spectrometry-based mass-to-charge value (m/z) yet of unknown structure. While crystallography and NMR spectroscopy undoubtedly have been of extraordinary importance in this process, their applicability in resolving metabolism’s fine structure has been restricted by their intrinsic requirement of sufficient and sufficiently pure materials. Unlike proteins that are made of fundamental amino acid building blocks, metabolites can be defined by the arrangement of the basic elements, typically C, H, N, O, P, and S, representing a virtually limitless array of chemical structures. This perspective describes this challenge, how it was originally addressed, and how metabolomics is evolving to address it today and in the future.
Introduction
Metabolite discovery has been fundamental to progress in biochemistry for three centuries. Urea, the first metabolite characterized by the 18th century scientist (and Elementa Chemiae author) Hermann Boerhaave, was possible largely due to its ubiquitous nature. Thus, setting the tone for the subsequent two centuries with identifications made solely on the most abundant metabolites (e.g., amino acids, sugars, and lipids). However, the finer details of metabolism started to emerge from the 1930’s onwards in parallel with the development of advanced and more sensitive analytical technologies, yielding a much higher resolution image of biochemistry. An image that has fundamentally altered our perceptions, constantly revealing the structural complexity and new metabolic activity within biochemistry.
Through these technological advances we have witnessed a rebirth of metabolism’s impact in every biological and therapeutic area, ranging from cancer, bioengineering, systems biology, the microbiome, and nutrition among numerous others. Metabolomics, understood as a compendium of technologies, has emerged to synergize with the classical biochemistry of the twentieth century and is facilitating further progress in biochemical research through the collective characterization and quantification of pools of metabolites that translate into the structure, function, and dynamics of an organism. For example, mass spectrometry technological developments, especially Nobel Prize winning electrospray ionization (ESI), allowed for the routine observation of intact molecular ions, a feat that was previously not possible for most biomolecules. ESI also enabled unimagined sensitivity that was orders of magnitude greater than their predecessors. More importantly, these technologies also allowed us to detect metabolites that were not previously known to exist in nature, or multicellular animals and plants, or unicellular microorganisms such as bacteria.
This last point, determining the structure of unknown metabolites, represents a major technological challenge and an opportunity to expand our biochemical knowledge base for both uncharacterized and well-characterized organisms. The goal to identifying all the known and unknown small molecules found within an organism, also known as the metabolome, represents the future of metabolism discoveries. Historically, from the 1700s to 1900s, any newly discovered metabolite built upon metabolism largely because of the ubiquitous nature of these metabolites. Ubiquitous often meant of fundamental importance and abundant enough to be isolated in optimal quantities to be characterized by the analytical techniques of that time (e.g., amino acids, monosaccharides, nucleotides, and so on). However, in the last decades, numerous new and less ubiquitous metabolites and classes of metabolites have been discovered, resulting in a sustained effort to characterize and decipher their physiological effects.
Soon after technological advancements allowed to decipher the fine structure of biochemical pathways and their physiological relevance, researchers started to target and leverage metabolism as a tool for drug development. For example, the birth control pill was developed in the 1950’s under the lead of Carl Djerassi (Watts, 2015). In the same period Bengt Samuelsson, Sune Bergström and John Vane started to decipher eicosanoid biology, founding the development of non-steroidal anti-inflammatory drugs and modern research fields such as inflammation resolution or the recently postulated inhibition of 15-PGDH for tissue regeneration (Bergstrom and Samuelsson, 1965; Buckley et al., 2014; Zhang et al., 2015). More recent examples of leveraging biochemical knowledge and bioactive metabolites as therapeutic strategies are the fatty acid esters of hydroxy fatty acids (FAHFAs) as anti-diabetic and anti-inflammatory lipids (Yore et al., 2014), the activation of the liver X receptors by the cholesterol precursor desmosterol in foaming macrophages (Spann et al., 2012), or the hijacking of cholesterol biosynthesis during hepatitis C virus infections (Rodgers et al., 2012). Other more diverse examples include oncometabolites as possible diagnostic markers and drug targets (Yang et al., 2013), the identification of the novel antibiotic teixobactin (Ling et al., 2015) as well as numerous classes of secondary plant metabolites such as alkaloids and flavonoids (Debnath et al., 2018). Together, these examples represent a fruitful knowledge base for metabolism research and drug discovery (Wishart, 2016).
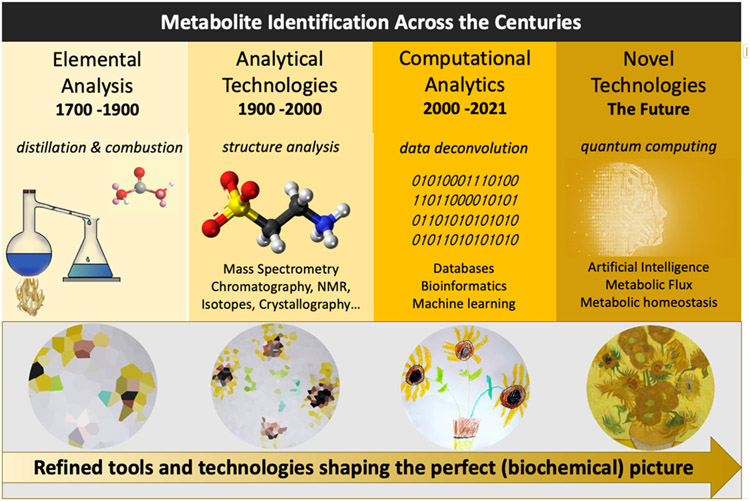
However, exciting new areas as for example the identification of microbiome derived metabolites with direct (patho-)physiological relevance (Nicholson et al., 2012) are emerging. While this has largely been established for short chain fatty acids, several new classes of metabolites are evolving with many yet to be discovered, discoveries that largely depend on metabolomic technologies (Han et al., 2021). This perspective describes the enduring obstacle of characterizing newly observed metabolites, how it was originally addressed, and how metabolomics is evolving to address it today and, in the future (Figure 1).
Figure 1.
A broad depiction of the evolution of metabolic sciences across the centuries as depicted by technology and an art metaphor (van Gogh). Over the centuries (bio)chemists have advanced their tools and skills, toward sketching the perfect biochemical picture. The years prior to the twentieth century largely focused on elemental analysis and scientific deduction of individual and purified chemical structures while the post-twentieth century saw significant steps forward in analytics. The twenty first century analysis of metabolism is represented by a confluence of analytical, computational, and artificial intelligence technologies towards the characterization of all metabolic constituents considered collectively.
1700s to 1900: The Beginning and Metabolite Characterization in Bulk
Take some very fresh well-concocted Urine of persons in perfect Health, put it preferentially into a very clean Vessel, and with an equable Heat of 200 degrees, evaporate it till you have reduced it to the consistence of fresh Cream” …“Put a large quantity of this thick inspissated Liquor into a tall cylindrical glass vessel with a paper tied over it and let it stand quite in a cool place for the space of a year…"
– Hermann Boerhaave (Duranton et al., 2016)
The first elemental composition determinations of small organic molecules such as urea, lactic acid, citric acid, or oxalic acid (Figure 2) were the result of applying analytical techniques developed by Boerhaave and Lavoisier in the 1700s which were later improved by Gay-Lussac and Thenard in the early 1800s. Their typical approach began with animal and food products particularly rich in specific molecules. For example, citric acid from lemon and lactic acid from fermented milk, followed by separating and purifying the constituents by distillation and crystallization, then deriving atomic weights by means of combustion analysis. While informative, these chemical formulas only allowed for structural hypothesis.
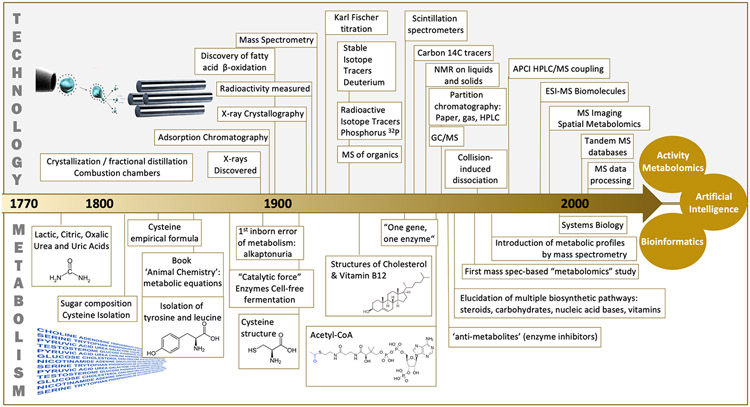
Figure 2.
A detailed view of how technological developments coincided with biochemical discoveries and the identification of previously unknown metabolites. Top image of electrospray ionization with a quadrupole mass spectrometer (Beadle and Tatum, 1941; Buchner, 1897; Chiewitz and Hevesy, 1935; Cooks, 1995; Ettre and Sakodynskii, 1993; Fischer, 1935; Garrod, 1902; Knoop, 1904; Purcell et al., 1946; Rabi et al., 1938; Röntgen, 1898; Woolley, 1959).
Throughout the nineteenth century the molecular formulas of many metabolites were determined (Thaulow, 1838), although the real breakthrough of this period was in establishing the basis of our knowledge of metabolic reactions, as recorded in a book by Justus von Liebig with the unlikely name “Animal Chemistry (Die Thier-Chemie)” (Freiherr von Liebig, 1843). Liebig inferred, for the first time, metabolic equations that described physiological processes without any evidence of the existence of such reactions in vivo, based solely on his knowledge of organic chemistry. Liebig's studies, therefore, laid the groundwork for analysis of the inter-conversions of simple organic molecules within the cell. Subsequently, the main methodological advance that would demonstrate Liebig's predicted metabolic reactions was the use of radioactive isotopes (3H, 32P, 14C). These same principles of “Animal Chemistry” as well as crystallography helped individuals like Louis Pasteur in their efforts to decipher metabolic structure and function. However, it should be emphasized that these efforts did not necessarily provide confirmed metabolite structures. This determination required a whole new set of technological advances introduced in the twentieth century. Nevertheless, outstanding intellectual achievements were made during this period, particularly when considering the available analytical technologies. Even today many physico-chemical techniques and properties are still used for substance characterization, including combustion analysis, boiling point (distillation) and melting point (capillary heating) determination, or specific chemical transformations. Many of these techniques are still an essential curricular part of chemistry/pharmacy studies and many of the reagents developed during the 1800s have evolved into widely accepted and applied color reactions; e.g., the Tollens test for reducing functional groups such as aldehydes (in reducing sugars) (Tollens, 1882) or the Marquis reaction which even today is a widely applied spot test for MDMA, phenylethylamines and opiates (Marquis, 1896). An example for the extraordinary commitment, effort and ingenuity needed to establish chemical structures during more than two centuries is the molecule cholesterol. Cholesterol, a 26-carbon, tetracyclic cyclopenta[a]phenanthrene essential to human life has first been discovered by François Poulletier de la Salle in gall stones presumably in 1758, however it took another half century until Michel-Eugène Chevreul pinpointed its physico-chemical properties and named it cholesterol (Chevreul, 1823; Schlienger, 2012). While Chevreul thoroughly characterized the substance, his studies yielded no significant insight into its actual chemical structure. In 1888, Friedrich Reinitzer established the exact molecular formula of cholesterol (Reinitzer, 1888, 1989), but it took until 1932 before the structure of cholesterol was correctly postulated chiefly based on the brilliant work of Heinrich O. Wieland and Adolf Windaus (Endo, 2010) (Butenandt, 1960). Finally, the more than 30 step total synthesis of cholesterol was published by the groups of Woodward and Robinson in 1951/52 (Mulheirn, 2000; Woodward et al., 1951), almost 200 hundred years after it was first discovered.
1900s: Analytical Technology Drives Discovery
The 1900s marked the beginning of numerous technological milestones that helped decipher many key biochemical processes in the central carbon metabolism of eukaryotic cells. Developments such as x-ray crystallography, the use of nuclear reactors as a source of artificial radioisotopes, and the development of scintillation spectrometers to replace Geiger counters (Rutherford and Geiger, 1908) produced exponential growth in biochemical research in the 1930s and beyond (Lipmann and Kaplan, 1946; Schoenheimer and Rittenberg, 1935; Windaus, 1932), including the discoveries of the TCA cycle by Hans Krebs (Buchanan, 2002; Manchester, 1998) following on previous contributions by Albert Györgi (Krebs, 1940), acetyl-CoA (Kresge et al., 2005a), glycolysis or the Embden-Meyerhof-Parnas pathway (Kresge et al., 2005b) as well as steroid biosynthesis. In that same period, chromatographic techniques developed by Archer Martin and Richard Synge (Martin and Synge, 1941) would quickly lead to methods such as gas chromatography and what would later be complemented with high-pressure liquid chromatography (HPLC). By 1945, virtually all the analytical techniques necessary for biochemical research were available to the next generation of researchers. In fact, by 1957, biosynthetic pathways for virtually all types of known biological molecules had already been elucidated, including lipids, carbohydrates, nucleic acid bases, amino acids, and vitamins. Most of this knowledge was compiled in 1955 by Donald Nicholson into a single map composed of about 20 metabolic pathways (Dagley and Nicholson, 1970). Therefore, many molecular formulas and metabolite structures had been discovered even before the first structure of a protein (myoglobin) with atomic resolution (Kendrew et al., 1958), the elucidation of the DNA structure in 1953, or the subsequent publication in 1958 of molecular biology’s central dogma. This situation would relegate the study of metabolism to a secondary effort in favor of the study of genes and proteins.
Nevertheless, the rise of nuclear magnetic resonance (NMR) and mass spectrometry (MS) boosted the development of advanced biochemical methods and metabolomics principles. NMR was first described by Isidor Rabi in 1938 and later used for the analysis of liquids and solids by Bloch and Purcell (Giunta and Mainz, 2020; Purcell et al., 1946; Rabi et al., 1938). The introduction of superconducting magnets during the 1970s, combined with Fourier transformation, rendered NMR applicable to the routine observation of 13C (the stable isotope of the carbon atom) in metabolic studies. Already in 1974, Seeley demonstrated the utility of NMR to detect metabolites in intact biological samples (Hoult et al., 1974). Mass spectrometry was first used to study organic molecules in 1934 by Conrad, although some of the most important advances to better contextualize what we know today as mass spectrometry-based metabolomics are due to Gohlke et al., McLafferty et al. and coupling gas chromatography to a mass spectrometer in 1959 (Gohlke, 1959), introducing the collision induced dissociation (CID) in 1968 (Haddon and McLafferty, 1968), and coupling liquid chromatography to mass spectrometry in 1974 (Arpino, 2006; Arpino et al., 1974).
The concept of individual biochemical profiles was developed in the late 1940s and early 1950s by Roger J. Williams (Williams, 1956). The first proof of concept of mass spectrometry-based metabolomics was described in 1966 by Dalgliesh et al. (Dalgliesh et al., 1966) when they carried out GC/MS experiment to separate and detect a wide range of metabolites present in urine and biological tissue extracts. Subsequently, Horning and colleagues introduced the term metabolic profiles, and together with Linus Pauling and Arthur Robinson, developed GC/MS methods to simultaneously monitor dozens of metabolites present in biological samples during the 1970s (Teranishi et al., 1972). Follow-up work by Gates and Sweeley further cemented the impact of GC/MS as a quantitative tool in metabolic profiling (Gates and Sweeley, 1978).
Even so, the cornerstone on which metabolomics (and proteomics) is mainly built was the development of electrospray ionization (ESI) for biomolecular analysis by John B. Fenn in 1989 (Fenn et al., 1989). Soon after, the first LC-ESI MS based untargeted metabolomics studies for the characterization of biological matrices were performed and the large potential of (untargeted) metabolite profiling was soon evident and appreciated (Cravatt et al., 1995; Lerner et al., 1994). These experiments also revealed key mass spectrometric improvements that needed to be addressed to effectively interpret untargeted mass spectral data and determine the structure of unknown metabolites, specifically the need for peak detection and alignment in convoluted LC/MS spectra for statistically characterizing meaningful metabolic features, and the need for tandem mass spectrometry databases for rapid identification.
During this same period, biopolymer analysis techniques applied to DNA and proteins marked the ascent of genomics in the 1980s and of proteomics in the 1990s. These cornerstone advancements captured great attention from the scientific community and laid the foundation for the future establishment of what is today known as systems biology. Soon, high-throughput genomic technologies became routine, which introduced the “big data” analysis problem. All these developments are now allowing for the convergence of the primary omics: genomics, proteomics, and metabolomics, a convergence made possible with the ultimate “omic-glue”: bioinformatics.
“The good into the pot, the bad into the crop” - The necessity of squeezing out the informative fraction of large data sets fueled the parallel development of bioinformatics and data mining pipelines. Some of these later efforts represented an excellent starting point for the analysis of information-rich datasets obtained in global metabolomics experiments. However, the genomics and proteomics era also stimulated the idea that the investigation of metabolites was a mature field, and that biochemistry was mainly driven by genes and proteins. Cellular metabolism was practically reduced to cellular processes by which nutrients were converted into energy metabolites, building blocks for the construction of DNA and proteins, and some other small organic end-products. In fact, most of the metabolic pathways that are taught today in the curricular program of biochemistry were discovered and mapped before 1960. This underlying concept influenced metabolic research over the last two decades of the 20th century. Residing within these boundaries, textbook metabolic pathways became a major target for biomarker discovery, fueled by technological advancements in LC-MS, including the development of ultra-performance liquid chromatography (UHPLC), translated into a multitude of methods for targeted metabolite quantification from well characterized metabolic pathways. As a natural consequence, targeted studies aimed at establishing metabolites as novel biomarkers of disease largely outnumbered other efforts to portray the components and functions of the metabolic machine.
21st Century Knowns: The Annotated Road to Known Metabolite Identification
As previously noted, identification of metabolites from the 1800s to 1900s was accomplished 'one at a time', that is, following purification and characterization of significant amounts of a single target compound from natural sources. In contrast, ‘omic’ technologies, and metabolomics, are comprehensive by definition, aiming at characterizing and quantifying all constituents considered collectively. However, the vast amount of data generated by modern, ultra-sensitive technologies is “curse and blessing”. On one hand it allows us to reshape and better define biochemistry, on the other hand, highly convoluted spectra resulting from adducts, isotopes, in-source fragments, and background and contaminant ions pose a significant challenge for metabolite identification. This is comparable to what has been described as “the cocktail party problem”, which is the task of hearing a sound of interest (i.e. sound segregation) masked by overlaying background noise (Woods and McDermott, 2018). This has led to a distinction between metabolite annotation and identification. While annotation refers to the assignment of a candidate metabolite to multiple and redundant MS signals based on analytical characteristics (e.g., retention time, m/z), identification is a much more tedious, nonetheless conclusive process assigning a chemical structure to a candidate metabolite. The latter either requires chemically pure standard materials for comparison or conclusive (2D) NMR data. Reporting standards for metabolite annotation and identification have been described by Salek et al. (Salek et al., 2013).
Consequently, the 21st century is characterized by increasingly sophisticated signal processing techniques for MS and NMR-based metabolomics allowing spectral annotations and (as far as possible) identifications from vast amounts of highly complex data. These include a multitude of peak detection and alignment software (Misra, 2021), e.g. XCMS (Smith et al., 2006), MZmine2 (Smith et al., 2006), Open-MS (Pfeuffer et al., 2017) and MS-DIAL (Tsugawa et al., 2015; Tsugawa et al., 2020) for LC/MS, or analog software for GC/MS such as eRah (Domingo-Almenara et al., 2016), ADAP-GC (Smirnov et al., 2019) or BinBase (Kind et al., 2009). Full-scan mass spectra information is complemented with tandem (MS/MS) mass spectral data for metabolite identification. MS/MS methods can produce structural information for hundreds or thousands of metabolites in minutes, are in constant evolution and improvement to match experimental MS/MS data with spectral databases. This also includes new mass spectral similarity scoring as a proxy for structural similarity (Huber et al., 2021). Moreover, as the employed analytical equipment in many cases influences the obtained data, individual large scale “in-house” databases for hundreds to thousands of metabolites with standardized analytical procedures are being established to facilitate metabolite identification and (relative) quantification rather than “simple” annotations. Such developments are made possible by the availability of large commercially available metabolite libraries (e.g. IROA technologies). Ultimately, (semi)-quantitative data and unambiguous metabolite identification are crucial for pathway analysis and integration with transcriptomics and genomics.
Still, spectral databases are a major component in the metabolite annotation process. For decades GC/MS has been the dominant identification technology primarily due to the impressive size of its chemical mass spectral libraries. For example, the National Institute for Standards and Technology (NIST) has a library of electron ionization (EI) mass spectra generated from over 300,000 individual compounds. However, as noted MS technologies have evolved dramatically since the advent of EI. The 2002 Nobel prizes highlighted this fact, awarding developments related to two ionization approaches that enabled the detection of intact biomolecules (ESI and soft laser desorption ionization). ESI has since become the dominant technology allowing for a broader range of molecules to be observed because it is “softer” (i.e., less destructive) and compatible with LC separation methods. Nonetheless, LC-ESI MS approaches initially suffered from the paucity of publicly available MS/MS spectra for small molecules and metabolites. This gap was filled in 2003 through the creation of the first database of ESI MS/MS spectra designed for the identification of small molecules and metabolites (Smith et al., 2005). Since 2003 numerous public and commercial databases and spectral libraries have been created, among these the METLIN (Xue et al., 2020) (Guijas et al., 2018) now with MS/MS experimental data on 860,000 molecular standards, Human Metabolome Database (Wishart et al., 2018), the Birmingham Metabolite Library Nuclear Magnetic Resonance database (Ludwig and Günther, 2011), BiGG (King et al., 2016)), MassBank (Horai et al., 2010), LipidMaps (Sud et al., 2007), mzCloud, the Fiehn lab GC-MS Database (Lai et al., 2018), and the Golm Metabolome Database (GMD) (Kopka et al., 2005).
Yet, advanced informatics approaches can pose a significant hurdle to the routine application of metabolomics strategies in non-specialized laboratories. Consequently, some of the main challenges in data analysis and informatics, have been addressed by the development of cloud-based technologies that integrate MS and MS/MS processes and tools into on-line platforms (Aron et al., 2020; Forsberg et al., 2018), but also commercial software and open access packages (e.g., OpenMS, MS-DIAL) (Röst et al., 2016; Tsugawa et al., 2015). Moreover, just recently workflow management systems have been introduced to metabolomics applications to facilitate reproducibility (Verhoeven et al., 2020).
21st Century Unknowns: The Road Less Traveled
An alternative path that most metabolomic scientists face on a regular basis, is trying to characterize unknown unknowns. The framework for the identification of unknown metabolites is more complex and directly connected to the many questions that remain to be addressed in metabolism research. How many metabolites compose the metabolome? What’s their origin and fate? What’s the functional role of these molecules in health and disease? To address these and many other intriguing inquiries, major technological advances are still required, with metabolite discovery (i.e., detection and annotation) and identification (i.e. structural elucidation) being certainly among the most urgent. Indeed, assigning identities to the tens of thousands of spectral signals from metabolome-wide studies is a time-consuming task, and the characterization of unknowns is a significant bottleneck. And to put it bluntly, biologists cannot afford to dedicate precious resources for biological research on wrongly assigned metabolic structures.
Unknown identification typically starts with the observation of a metabolic feature(s) dysregulated over distinct experimental conditions (e.g., heath vs disease, wildtype vs mutant, etc.) for which no putative identity is available. First, the accurate m/z measurement for the molecular ion is used to compute a set of molecular formulas that are compatible with the detected mass. The technical improvements in MS accuracy achieved over the last two decades are of crucial importance at this stage, as these enable narrowing down the number of possible candidates, from tens of thousands to tens of features. Moreover, bioinformatic tools such as enviPat (Loos et al., 2015) can be used to rapidly predict the isotopic pattern for each formula: all those patterns not consistent with the experimental data can be excluded from the list of candidates. Further information can be obtained from the presence of adducts, neutral losses, in-source fragments (Guijas et al., 2018) and from the analysis of homologous series of features (Loos and Singer, 2017) in the MS data, the latter being particularly informative for the characterization of lipids. The unknown feature is subsequently fragmented at different collision energies, and the MS/MS spectra manually interpreted with the aid of bioinformatic tools, including, for instance, the fragment similarity as well as neutral loss analysis. Additional evidence can be gathered from MS/MS spectra reported in bibliography and in data-driven social-network repositories for MS/MS data sharing and curation, such as the GNPS (Wang et al., 2016), and, when possible, through the analysis of MS/MS data from stable isotope-labeling experiments (Mahieu et al., 2014). Complementary to MS, new NMR-based approaches allow for de novo identification of unknown molecular structures in complex mixtures, without the need for extensive purification (Emwas et al., 2019). Yet, inferred molecular structures still need to be confirmed by the analysis of pure standards obtained via chemical synthesis (Kalisiak et al., 2009). A specific case has just recently been made for lipids. Using a decision tree approach the identification of known and unknowns could be accomplished across several analytical platforms (Hartler et al., 2017).
21st Century: The Road Ahead
The beginning of the 21st century saw significant progress in large-scale genomic, transcriptomic and proteomic approaches (Malmström et al., 2007) as a result of new technologies and bioinformatic tools that allowed for the amplification and subsequent accurate characterization of the sequence of monomers in DNA/RNA and proteins, namely, nucleotides and amino acids, respectively. Metabolites, in contrast, are not sequences of monomers and do not result from a residue-by-residue transfer of information. Therefore, metabolomics trailed these genomic and proteomic developments with few novel biochemical connections being drawn on the metabolic map, as metabolic research relied deeply on a priori knowledge. Yet, over the last two decades, improved application of LC-MS/MS coupled with bioinformatics paved the way for metabolome-wide investigations that moved research from targeted studies to more comprehensive analyses, enabling the discovery of novel metabolites and the occurrence of metabolites previously unknown in certain organisms, thereby assisting gene function annotations. Key improvements in the analytical hardware included higher speed (i.e. scan capabilities), selectivity (i.e. resolution and mass accuracy) and sensitivity (i.e. signal intensity), which has increased metabolome coverages despite the inherent challenges due to their large dynamic range (>5 order-of-magnitude differences in the concentrations of endogenous metabolites). Nonetheless, the availability of a comprehensive database of tandem mass spectra for exogenous and endogenous metabolites has been a bottleneck in streamlining a robust workflow in metabolomics and has consequently been addressed by several initiatives.
The multi-pronged effort in the creation of empirical MS/MS databases from pure standards represent a significant step towards the automated identification of known metabolites; however, even now these resources cover less than 1% of the known small molecule space (Xue et al., 2020). Equally important, the biochemical landscape of prokaryotic and eukaryotic organisms within public and commercial mass spectral databases, average only 40% of nodes in metabolic networks (Frainay et al., 2018). However, with databases growing at a rapid pace, the hope is that many of the “known unknowns” will be covered in the coming decades. Part of this optimism lies in using existing data for training the next generation of metabolite identification tools based on Artificial Intelligence (AI) (Nguyen et al., 2019). In silico methods offer a possible solution to the problem of incomplete spectral libraries and inadequate collections of compounds, reducing the manual effort required for the assignment of the MS/MS signals to molecular substructures (i.e., fragments), diminishing misidentified metabolites and helping to distinguish closely related substances (McEachran et al., 2019). Yet, many obstacles for successful database matching remain, e.g., very low abundant metabolites, a lack of synthetic standard materials, or structurally highly similar substances such as geometric isomers and enantiomers.
However, in silico approaches are gaining traction in the metabolomics field and since the appearance of AI at the end of the first half of the 20th century, the technology has evolved greatly. In the 1980s, a branch of AI was born: machine learning (ML). ML is an analytical way of solving problems through identification, classification, or prediction. ML can gather and build knowledge from complex datasets (e.g., MS/MS data from metabolite repositories), that can be leveraged to generate predictive rules for interpretating experimental MS/MS data and generating structural hypothesis in the de novo identification of unknowns (Aguilar-Mogas et al., 2017). ML methods can also learn intermediate representations, such as molecular fingerprints (Cereto-Massagué et al., 2015) from historical spectrum–structure relationships (Dührkop et al., 2015). For example, Liu et al. have recently provided a proof of concept using an association rule mining strategy for metabolite substructure auto-recommendation (MESSAR) (Liu et al., 2020). The embedment of this type of tools in the untargeted metabolomics pipeline will significantly ease/facilitate the interpretation of the MS/MS data, especially if trained on comprehensive databases such as METLIN, NIST, MassBank or mzCloud. Already in 2011, a branch of machine learning called deep learning (DL) appeared. While ML operates with regression algorithms or decision trees (e.g., random forest), DL uses neural networks that function very similar to biological neural connections of our brain. DL can also be used to target compounds for which neither spectral nor structural reference data are available and predict classes lacking tandem mass spectrometry training sets (Dührkop et al., 2021).
Other AI related branches such as natural language processing (NLP), hold the potential for annotating molecules for which no reference spectra exist and to expose biochemical relationships between molecules (van der Hooft et al., 2016). In other words, embedding metabolic features within their relevant biological context under specific experimental conditions, thereby increasing confidence annotations and shortening putative candidate lists (Majumder et al., 2021). Such applications implement cognitive literature mining to sketch relations with bibliographic records. For instance, through a deep exploration of more than 300,000 abstracts, Warth et al. recently showcased the utility of cognitive computing for prioritizing metabolite annotations from global exposomics, enabling comprehensive and rapid exposure assessment (Warth et al., 2017). Cognitive computing is generally described as using NLP and ML to extract key concepts from the scientific literature, understand the semantic context, and predict and identify potential connections between entities not explicitly described in the text.
Although, this view of the predictive algorithms may be overly optimistic, especially when one considers the challenge at hand, which is identifying a complex chemical structure from a relatively small set of numeric variables: m/z values of precursor and a few fragment ions. The major challenge lies in not dealing with a convenient set of building blocks (e.g., amino acids for proteins) such as those produced by direct cleavage of peptide bonds in proteomic MS/MS experiments, and instead an almost endless number of arrangements that can occur to create metabolites. A prominent example is the very biologically active yet difficult to characterize oxylipins (Galano et al., 2017; Kloos et al., 2014). So, for example, the hope that one could use AI as a tool to use MS/MS data to decipher the complex structure of certain molecules is akin to asking AI to decipher the structure of a Swiss watch after it has been blown apart.
Given a metabolite’s complexity, likely it will come down to empirical data to solve their structures, especially since a biologist should not commit to investigating a molecule’s function (often requiring years of effort) based purely on bioinformatic conjecture. The candidate structure obtained from the analysis of MS/MS data must undergo confirmation using standards and orthogonal analytical techniques, for unequivocal identification. NMR is currently considered the method of choice to accomplish this task; however, it has relatively low sensitivity. This limits its applicability to the identification of abundant metabolites in biological samples or to the confirmation of the hypothesized structure using standards obtained through chemical synthesis.
Mass Spectrometry, NMR, and Orthogonal Technologies
Emerging technologies will likely play a key role in the future of metabolite identification. For example, cryogenic electron microscopy (cryo-EM) has shown impressive results with proteins and macromolecular complexes and has recently demonstrated promise for the direct characterization of small molecules (Jones et al., 2018; Scapin et al., 2018). Additionally, geometric and stereochemical considerations such as double geometry or chiral centers remain elusive to most tandem mass spectrometry-based identification approaches and will likely take center stage in future investigations involving differential ion mobility MS approaches targeting the chiral space of the metabolome. Novel fragmentation techniques such as electron-activated dissociation (EAD) (Baba et al., 2021), OZid (Thomas et al., 2008) and EIEIO (Baba et al., 2018) are technologies that are evolving as useful additions. Although, some successes have been accomplished for separating geometric isomers and enantiomers using ion mobility-based techniques (Jónasdóttir et al., 2015; Xie et al., 2021) these approaches are far from being routine. Yet, oncometabolites (e.g. 2R-hydroxyglutarte (Bunse et al., 2018)) or even simple D-lactic acid (the metabolic culprit for D-lactic acidosis (Fabian et al., 2017)) underline the importance of further deciphering the chiral fine structure of metabolism. The combination of novel fragmentation techniques and MSn will likely facilitate these identifications, although not in itself, but rather through the accumulation of large amounts of data on many molecules, following deconvolution using AI methods.
Next to MS based technologies, advancements in separation technologies are still ongoing (Kohler and Giera, 2017), with for example the revival of supercritical fluid chromatography (van de Velde et al., 2020) or advanced LC separations starting to challenge GC based technologies with respect to separation efficiency (Plumb et al., 2021). These developments will further advance metabolite identification, helping to deconvolute MS data and overcome matrix effects.
In addition to MS based approaches, NMR can still be considered the gold standard for metabolite identification, given sufficient and sufficiently pure material is available. However, NMR has become established for the quantitative analysis of cellular energy metabolism (Kostidis et al., 2017), the analysis of lipoproteins including subfractions (Lodge et al., 2021), metabolic phenotyping (Beckonert et al., 2007) as well as for isotopic flux and tracing experiments (Nagana Gowda and Raftery, 2021). Moreover, new developments such as for example isotopic tags or micro-coil NMR aim at increasing sensitivity and hence metabolic coverage of NMR based applications (Nagana Gowda and Raftery, 2015).
To this point and beyond, novel analytical technologies and the application of innovative techniques of mass spectrometry for metabolome-wide studies and autonomous bioinformatic pipelines coupled with comprehensive spectral databases triggered a paradigm shift in biology and other life sciences: from studies driven by a priori hypothesis to unbiased global investigations that had little or no a priori considerations (Carroll and Goodstein, 2009). This change of perspective also highlighted, surprisingly, the presence of known and unknown molecules correlating with specific biological conditions/phenotypes, pointing to the existence of a metabolic “black matter” yet to be discovered. For example, Vizcaino et al. recently used a global metabolomics approach for pinpointing a group of small molecules named colibactins from E. coli implicated in colon cancer (Vizcaino and Crawford, 2015; Vizcaino et al., 2014). The existence of these metabolites was first inferred from the observation of a specific genomic cluster (pks or clb) encoding for a hybrid polyketide synthase (PKS)/non-ribosomal peptide synthase (NRPS) biosynthetic system in the bacterial strains correlating with tumor growth. It was thus postulated that these bacteria facilitate cancer through the release of metabolites that cause chromosome instability and DNA damage. Comparative metabolomics analysis of mutants obtained by deleting the clb cluster vs the wildtype bacteria enabled the isolation of a set of molecules, which structure was further characterized by targeted MS/MS experiments and isotope labeling studies. Finally, the structures were confirmed by NMR and by chemical synthesis, and their genotoxic activity validated in vitro (Shine et al., 2018) and through the direct detection of colibactin-DNA adducts in colonic epithelial cells of mice monocolonized with pks+ E. coli (Wilson et al., 2019). Nevertheless, due to its instability and highly complex structure, only a multi-disciplinary approach did finally lead to a concise biosynthetic route as well as a description of its structure and its DNA cross-links (Xue et al., 2019). However, natural colibactin has to date eluted isolation and many aspects of its bioactivities and particularly of its metabolites are still under discussion (Carlson and Balskus, 2019; Herzon, 2020).
Ultimately, the metabolic black matter hypothesis has been further supported by genome-wide studies (Shin et al., 2014) in conjunction with bioinformatic predictions (Donia et al., 2014) and isotope tracing experiments (Zamboni et al., 2015). Although, we cannot yet precisely establish how many unknowns remain to be characterized, it is evident that new technologies and metabolomic approaches have already discovered new metabolites, metabolic reactions and unexpected metabolic fluxes that have important physiological relevance (Bowen and Northen, 2010). This also applies to the actual direction of specific enzymatic reactions and stereoisomers.
Today, stable isotope labeling studies can provide further insight for understanding the biosynthetic origin and fate of knowns and unknowns (Huang et al., 2014). Stable isotopes, such as 13C and 15N, are much safer and informative alternatives to the formerly used radioactive isotopes. The recent development of bioinformatic tools for labeled data processing (e.g., the X13CMS, geoRge), and the creation of a database serving stable isotope-based metabolomics (isoMETLIN) significantly expedited the data analysis and interpretation pipeline (Capellades et al., 2016; Cho et al., 2014). For example, in 2016 we applied a global isotope labeling approach and the X13CMS package to disentangle the contribution from the well-established ammonium metabolic pathway as opposed to the alternative nitrate assimilation route in nitrate reducing bacteria. The cultures were fully labeled using 15NO3 as nitrogen source, and subsequently analyzed by global metabolomics. This study led to the unexpected finding that certain strains of nitrate reducing bacteria can adopt survival strategies based on the co-utilization of nitrate and ammonia (Kurczy et al., 2016). Other highly relevant studies include organ specific metabolic flux studied by infusing carbon labelled nutrients (Hui et al., 2020) as well as metabolite exchange between mammalian organs (Jang et al., 2019). Ultimately, metabolic isotope tracing and flux analysis are crucial to sketch a detailed picture of healthy and diseased metabolism opening new therapeutic and diagnostic applications (Jang et al., 2018). Likely, isotope tracing and flux will next be tackled on-tissue (organoids) enabled by the rise of advanced mass spectrometry imaging applications.
Taken together, the LC, MS and NMR, and bioinformatic advancements made over the last two decades of this century enabled numerous insights and demonstrated global metabolomics as a powerful discovery technology for biochemical studies. Together they also represent part of the future for metabolite identification of unknowns, although it will likely be a multi-component effort that will include MS, NMR, ion mobility, AI, cryoEM, as well as synthetic chemical methods. Future efforts for streamlining metabolic research will likely revolve around five main goals: i) developing more reliable autonomous untargeted metabolomic data analysis platforms to pull out differentially regulated features, ii) the further inclusion of experimental MS/MS data for known unknowns in molecular databases for the autonomous identification of known metabolites, iii) the automated generation of structural hypothesis for deciphering unknown unknowns, including stereoisomers, iv) the development of solutions for pathway analysis, and v) prioritization approaches for identifying metabolites to test for biological activity.
Conclusion
Urea represented the first puzzle piece in characterizing the mosaic that is metabolism, a mosaic that continues to grow even to this day. Whether it is the simplest of structures (e.g., urea), the more complex acetyl-CoA, or brand-new metabolites, these discoveries gradually provide us with a more complete mechanistic understanding. The metabolomics era - the youngest of the primary omics, driving these discoveries can be characterized by a combination of analytical and computational advances enabling the characterization of metabolites and their respective metabolism. These technologies are providing a more complete understanding of metabolism, yet more intriguing, is how we can use these discoveries to drive biological activity within any given system. These technologies will greatly accelerate the workflow for the characterization of the numerous metabolic unknowns detected in global metabolomics data. However, we believe that this stage will be just a step towards the accomplishment of a more exciting goal, which lies in the comprehensive kinetic definition of metabolism as well as the discovery of the biological activity of metabolites in health and disease, through “Activity Metabolomics” (Jang et al., 2018; Rinschen et al., 2019). The emergence of activity metabolomics is already showing us the central dogma of molecular biology needs to be reconsidered as metabolites are recognized for their true value, master manipulators of biology.
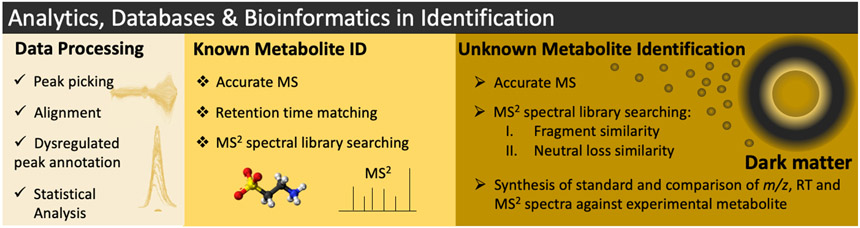
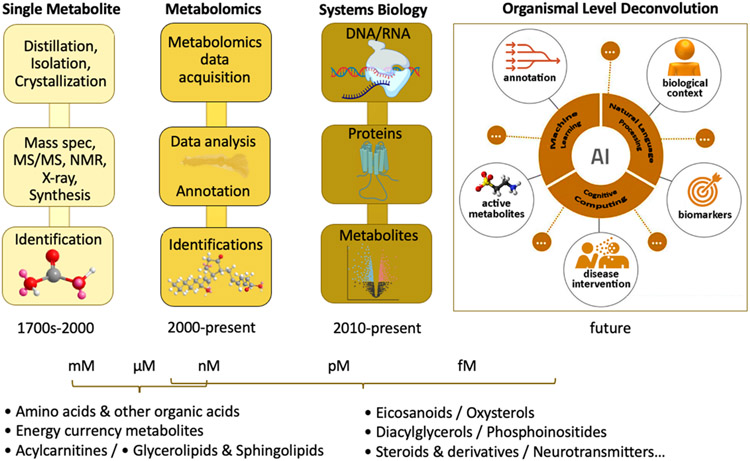
Figure 3.
Novel signal processing techniques for MS and NMR allow peak detection, alignment, deconvolution, and spectral matching via MS/MS databases of standards, and statistical assessment of metabolomic data followed by known and unknown metabolite identification. The timeline for unknown identification can vary from days to years depending on the complexity of the chemical structure. Also, it depends on the complexity of the synthesis and the amount of biological material available.
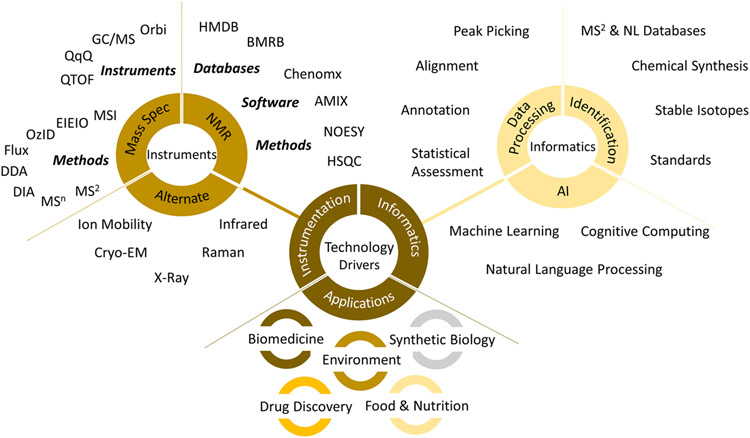
Figure 4.
A compendium of technologies that illustrate the state-of-the-art.
Figure 5.
Cognitive computing and AI are altering the way big data are processed and integrated. New natural language processing (NLP) platforms are emerging for biologists in other fields and in metabolomics. NLP provides literature-based contextualization of spectral and metabolic features that decreases the time and expert-level subject knowledge required during the prioritization, identification, and interpretation steps in the metabolomics data analysis pipeline.
References
- Aguilar-Mogas A, Sales-Pardo M, Navarro M, Guimerà R, and Yanes O (2017). iMet: A Network-Based Computational Tool To Assist in the Annotation of Metabolites from Tandem Mass Spectra. Analytical Chemistry 89, 3474–3482. [DOI] [PubMed] [Google Scholar]
- Aron AT, Gentry EC, McPhail KL, Nothias L-F, Nothias-Esposito M, Bouslimani A, Petras D, Gauglitz JM, Sikora N, Vargas F, et al. (2020). Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nature Protocols 15, 1954–1991. [DOI] [PubMed] [Google Scholar]
- Arpino P, ed. (2006). History of LC-MS development and interfacing. (Elsevier; ). [Google Scholar]
- Arpino P, Baldwin MA, and McLafferty FW (1974). Liquid chromatography-mass spectrometry. II—continuous monitoring. Biomedical Mass Spectrometry 1, 80–82. [DOI] [PubMed] [Google Scholar]
- Baba T, Campbell JL, Le Blanc JCY, Baker PRS, and Ikeda K (2018). Quantitative structural multiclass lipidomics using differential mobility: electron impact excitation of ions from organics (EIEIO) mass spectrometry. Journal of lipid research 59, 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ryumin P, Duchoslav E, Chen K, Chelur A, Loyd B, and Chernushevich I (2021). Dissociation of Biomolecules by an Intense Low-Energy Electron Beam in a High Sensitivity Time-of-Flight Mass Spectrometer. Journal of the American Society for Mass Spectrometry. [DOI] [PubMed] [Google Scholar]
- Beadle GW, and Tatum EL (1941). Genetic Control of Biochemical Reactions in Neurospora. Proceedings of the National Academy of Sciences 27, 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckonert O, Keun HC, Ebbels TM, Bundy J, Holmes E, Lindon JC, and Nicholson JK (2007). Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nature Protocols 2, 2692–2703. [DOI] [PubMed] [Google Scholar]
- Bergstrom S, and Samuelsson B (1965). Prostaglandins. Annual Review of Biochemistry 34, 101–108. [DOI] [PubMed] [Google Scholar]
- Bowen BP, and Northen TR (2010). Dealing with the unknown: metabolomics and metabolite atlases. Journal of the American Society for Mass Spectrometry 21, 1471–1476. [DOI] [PubMed] [Google Scholar]
- Buchanan JM (2002). Biochemistry during the Life and Times of Hans Krebs and Fritz Lipmann. Journal of Biological Chemistry 277, 33531–33536. [DOI] [PubMed] [Google Scholar]
- Buchner E (1897). Alkoholische Gährung ohne Hefezellen. Berichte der deutschen chemischen Gesellschaft 30, 117–124. [Google Scholar]
- Buckley CD, Gilroy DW, and Serhan CN (2014). Proresolving lipid mediators and mechanisms in the resolution of acute inflammation. Immunity 40, 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunse L, Pusch S, Bunse T, Sahm F, Sanghvi K, Friedrich M, Alansary D, Sonner JK, Green E, Deumelandt K, et al. (2018). Suppression of antitumor T cell immunity by the oncometabolite (R)-2-hydroxyglutarate. Nature Medicine 24, 1192–1203. [DOI] [PubMed] [Google Scholar]
- Butenandt A (1960). Zur Geschichte der Sterin- und Vitamin-Forschung. Adolf Windaus zum Gedächtnis. Angewandte Chemie 72, 645–651. [Google Scholar]
- Capellades J, Navarro M, Samino S, Garcia-Ramirez M, Hernandez C, Simo R, Vinaixa M, and Yanes O (2016). geoRge: A Computational Tool To Detect the Presence of Stable Isotope Labeling in LC/MS-Based Untargeted Metabolomics. Analytical Chemistry 88, 621–628. [DOI] [PubMed] [Google Scholar]
- Carlson ES, and Balskus EP (2019). The mysteries of macrocyclic colibactins. Nature Chemistry 11, 867–869. [DOI] [PubMed] [Google Scholar]
- Carroll S, and Goodstein D (2009). Defining the scientific method. Nature methods 6, 237–237. [DOI] [PubMed] [Google Scholar]
- Cereto-Massagué A, Ojeda MJ, Valls C, Mulero M, Garcia-Vallvé S, and Pujadas G (2015). Molecular fingerprint similarity search in virtual screening. Methods (San Diego, Calif.) 71, 58–63. [DOI] [PubMed] [Google Scholar]
- Chevreul ME (1823). A chemical study of oils and fats of animal origin. (St Eutrope-de-Born: Sàrl Dijkstra-Tucker; ). [Google Scholar]
- Chiewitz O, and Hevesy G (1935). Radioactive Indicators in the Study of Phosphorus Metabolism in Rats. Nature 136, 754–755. [Google Scholar]
- Cho K, Mahieu N, Ivanisevic J, Uritboonthai W, Chen Y Jr., Siuzdak G, and Patti GJ (2014). isoMETLIN: A Database for Isotope-Based Metabolomics. Analytical Chemistry 86, 9358–9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooks RG (1995). Special feature: Historical. Collision-induced dissociation: Readings and commentary. Journal of Mass Spectrometry 30, 1215–1221. [Google Scholar]
- Cravatt BF, Prospero-Garcia O, Siuzdak G, Gilula NB, Henriksen SJ, Boger DL, and Lerner RA (1995). Chemical characterization of a family of brain lipids that induce sleep. Science (New York, N.Y.) 268, 1506–1509. [DOI] [PubMed] [Google Scholar]
- Dagley S, and Nicholson DE (1970). An Introduction to Metabolic Pathways. (Blackwell Scientific Publications Ltd.). [Google Scholar]
- Dalgliesh CE, Horning EC, Horning MG, Knox KL, and Yarger K (1966). A gas-liquid-chromatographic procedure for separating a wide range of metabolites occuring in urine or tissue extracts. Biochem J 101, 792–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath B, Singh WS, Das M, Goswami S, Singh MK, Maiti D, and Manna K (2018). Role of plant alkaloids on human health: A review of biological activities. Materials Today Chemistry 9, 56–72. [Google Scholar]
- Domingo-Almenara X, Brezmes J, Vinaixa M, Samino S, Ramirez N, Ramon-Krauel M, Lerin C, Díaz M, Ibáñez L, Correig X, et al. (2016). eRah: A Computational Tool Integrating Spectral Deconvolution and Alignment with Quantification and Identification of Metabolites in GC/MS-Based Metabolomics. Analytical Chemistry 88, 9821–9829. [DOI] [PubMed] [Google Scholar]
- Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, Clardy J, Linington RG, and Fischbach MA (2014). A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158, 1402–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dührkop K, Nothias L-F, Fleischauer M, Reher R, Ludwig M, Hoffmann MA, Petras D, Gerwick WH, Rousu J, Dorrestein PC, et al. (2021). Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nature biotechnology 39, 462–471. [DOI] [PubMed] [Google Scholar]
- Dührkop K, Shen H, Meusel M, Rousu J, and Böcker S (2015). Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proceedings of the National Academy of Sciences 112, 12580–12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duranton F, Jankowski J, Więcek A, and Argilés À (2016). On the discovery of UREA. Identification, synthesis and observations that let to establishing the first uraemic retention solute. Giornale italiano di nefrologia : organo ufficiale della Societa italiana di nefrologia 33 Suppl 66, 33.s66.16. [PubMed] [Google Scholar]
- Emwas A-H, Roy R, McKay RT, Tenori L, Saccenti E, Gowda GAN, Raftery D, Alahmari F, Jaremko L, Jaremko M, et al. (2019). NMR Spectroscopy for Metabolomics Research. Metabolites 9, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo A (2010). A historical perspective on the discovery of statins. Proceedings of the Japan Academy. Series B, Physical and biological sciences 86, 484–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettre LS, and Sakodynskii KI (1993). M. S. Tswett and the discovery of chromatography II: Completion of the development of chromatography (1903–1910). Chromatographia 35, 329–338. [Google Scholar]
- Fabian E, Kramer L, Siebert F, Högenauer C, Raggam RB, Wenzl H, and Krejs GJ (2017). D-lactic acidosis - case report and review of the literature. Zeitschrift fur Gastroenterologie 55, 75–82. [DOI] [PubMed] [Google Scholar]
- Fenn J, Mann M, Meng C, Wong S, and Whitehouse C (1989). Electrospray ionization for mass spectrometry of large biomolecules. Science (New York, N.Y.) 246, 64–71. [DOI] [PubMed] [Google Scholar]
- Fischer K (1935). Neues Verfahren zur maßanalytischen Bestimmung des Wassergehaltes von Flüssigkeiten und festen Körpern. Angewandte Chemie 48, 394–396. [Google Scholar]
- Forsberg EM, Huan T, Rinehart D, Benton HP, Warth B, Hilmers B, and Siuzdak G (2018). Data processing, multi-omic pathway mapping, and metabolite activity analysis using XCMS Online. Nature Protocols 13, 633–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frainay C, Schymanski EL, Neumann S, Merlet B, Salek RM, Jourdan F, and Yanes O (2018). Mind the Gap: Mapping Mass Spectral Databases in Genome-Scale Metabolic Networks Reveals Poorly Covered Areas. Metabolites 8, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiherr von Liebig J (1843). Die Thier-Chemie, oder, Die organische Chemie in ihrer Anwendung auf Physiologie und Pathologie. (Braunschweig: F. Vieweg und Sohn; ). [Google Scholar]
- Galano JM, Lee YY, Oger C, Vigor C, Vercauteren J, Durand T, Giera M, and Lee JC (2017). Isoprostanes, neuroprostanes and phytoprostanes: An overview of 25years of research in chemistry and biology. Progress in lipid research 68, 83–108. [DOI] [PubMed] [Google Scholar]
- Garrod A (1902). THE INCIDENCE OF ALKAPTONURIA : A STUDY IN CHEMICAL INDIVIDUALITY. The Lancet 160, 1616–1620. [Google Scholar]
- Gates SC, and Sweeley CC (1978). Quantitative metabolic profiling based on gas chromatography. Clinical Chemistry 24, 1663–1673. [PubMed] [Google Scholar]
- Giunta CJ, and Mainz VV (2020). Discovery of Nuclear Magnetic Resonance: Rabi, Purcell, and Bloch. In Pioneers of Magnetic Resonance (American Chemical Society), pp. 3–20. [Google Scholar]
- Gohlke RS (1959). Time-of-Flight Mass Spectrometry and Gas-Liquid Partition Chromatography. Analytical Chemistry 31, 535–541. [Google Scholar]
- Guijas C, Montenegro-Burke JR, Domingo-Almenara X, Palermo A, Warth B, Hermann G, Koellensperger G, Huan T, Uritboonthai W, Aisporna AE, et al. (2018). METLIN: A Technology Platform for Identifying Knowns and Unknowns. Analytical Chemistry 90, 3156–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddon WF, and McLafferty FW (1968). Metastable ion characteristics. VII. Collision-induced metastables. Journal of the American Chemical Society 90, 4745–4746. [Google Scholar]
- Han S, Van Treuren W, Fischer CR, Merrill BD, DeFelice BC, Sanchez JM, Higginbottom SK, Guthrie L, Fall LA, Dodd D, et al. (2021). A metabolomics pipeline for the mechanistic interrogation of the gut microbiome. Nature 595, 415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartler J, Triebl A, Ziegl A, Trötzmüller M, Rechberger GN, Zeleznik OA, Zierler KA, Torta F, Cazenave-Gassiot A, Wenk MR, et al. (2017). Deciphering lipid structures based on platform-independent decision rules. Nature methods 14, 1171–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzon SB (2020). Macrocyclic colibactins. Nature Chemistry 12, 1005–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, et al. (2010). MassBank: a public repository for sharing mass spectral data for life sciences. Journal of mass spectrometry 45, 703–714. [DOI] [PubMed] [Google Scholar]
- Hoult DI, Busby SJW, Gadian DG, Radda GK, Richards RE, and Seeley PJ (1974). Observation of tissue metabolites using 31P nuclear magnetic resonance. Nature 252, 285–287. [DOI] [PubMed] [Google Scholar]
- Huang X, Chen Y Jr., Cho K, Nikolskiy I, Crawford PA, and Patti GJ (2014). X13CMS: Global Tracking of Isotopic Labels in Untargeted Metabolomics. Analytical Chemistry 86, 1632–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber F, Ridder L, Verhoeven S, Spaaks JH, Diblen F, Rogers S, and van der Hooft JJJ (2021). Spec2Vec: Improved mass spectral similarity scoring through learning of structural relationships. PLOS Computational Biology 17, e1008724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui S, Cowan AJ, Zeng X, Yang L, TeSlaa T, Li X, Bartman C, Zhang Z, Jang C, Wang L, et al. (2020). Quantitative Fluxomics of Circulating Metabolites. Cell Metabolism 32, 676–688.e674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang C, Chen L, and Rabinowitz JD (2018). Metabolomics and Isotope Tracing. Cell 173, 822–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang C, Hui S, Zeng X, Cowan AJ, Wang L, Chen L, Morscher RJ, Reyes J, Frezza C, Hwang HY, et al. (2019). Metabolite Exchange between Mammalian Organs Quantified in Pigs. Cell Metabolism 30, 594–606.e593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jónasdóttir HS, Papan C, Fabritz S, Balas L, Durand T, Hardardottir I, Freysdottir J, and Giera M (2015). Differential mobility separation of leukotrienes and protectins. Analytical Chemistry 87, 5036–5040. [DOI] [PubMed] [Google Scholar]
- Jones CG, Martynowycz MW, Hattne J, Fulton TJ, Stoltz BM, Rodriguez JA, Nelson HM, and Gonen T (2018). The CryoEM Method MicroED as a Powerful Tool for Small Molecule Structure Determination. ACS Central Science 4, 1587–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalisiak J, Trauger SA, Kalisiak E, Morita H, Fokin VV, Adams MWW, Sharpless KB, and Siuzdak G (2009). Identification of a New Endogenous Metabolite and the Characterization of Its Protein Interactions through an Immobilization Approach. Journal of the American Chemical Society 131, 378–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrew JC, Bodo G, Dintzis HM, Parrish RG, Wyckoff H, and Phillips DC (1958). A three-dimensional model of the myoglobin molecule obtained by x-ray analysis. Nature 181, 662–666. [DOI] [PubMed] [Google Scholar]
- Kind T, Wohlgemuth G, Lee DY, Lu Y, Palazoglu M, Shahbaz S, and Fiehn O (2009). FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Analyical Chemistry 81, 10038–10048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King ZA, Lu J, Dräger A, Miller P, Federowicz S, Lerman JA, Ebrahim A, Palsson BO, and Lewis NE (2016). BiGG Models: A platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res 44, D515–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloos D, Lingeman H, Mayboroda OA, Deelder AM, Niessen WMA, and Giera M (2014). Analysis of biologically-active, endogenous carboxylic acids based on chromatography-mass spectrometry. TrAC Trends in Analytical Chemistry 61, 17–28. [Google Scholar]
- Knoop F (1904). Der Abbau aromatischer fettsäuren im tierkörper. Beitraege zur Chemischen Physiologie und Pathologie 6, 150–162. [Google Scholar]
- Kohler I, and Giera M (2017). Recent advances in liquid-phase separations for clinical metabolomics. Journal of Separation Science 40, 93–108. [DOI] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, et al. (2005). GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics (Oxford, England) 21, 1635–1638. [DOI] [PubMed] [Google Scholar]
- Kostidis S, Addie RD, Morreau H, Mayboroda OA, and Giera M (2017). Quantitative NMR analysis of intra- and extracellular metabolism of mammalian cells: A tutorial. Analytica chimica acta 980, 1–24. [DOI] [PubMed] [Google Scholar]
- Krebs HA (1940). The citric acid cycle and the Szent-Györgyi cycle in pigeon breast muscle. Biochem J 34, 775–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresge N, Simoni RD, and Hill RL (2005a). Fritz Lipmann and the Discovery of Coenzyme A. Journal of Biological Chemistry 280, 164–166.15507456 [Google Scholar]
- Kresge N, Simoni RD, and Hill RL (2005b). Otto Fritz Meyerhof and the Elucidation of the Glycolytic Pathway. Journal of Biological Chemistry 280, e3. [PubMed] [Google Scholar]
- Kurczy ME, Forsberg EM, Thorgersen MP, Poole FL 2nd, Benton HP, Ivanisevic J, Tran ML, Wall JD, Elias DA, Adams MW, et al. (2016). Global Isotope Metabolomics Reveals Adaptive Strategies for Nitrogen Assimilation. ACS chemical biology 11, 1677–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Tsugawa H, Wohlgemuth G, Mehta S, Mueller M, Zheng Y, Ogiwara A, Meissen J, Showalter M, Takeuchi K, et al. (2018). Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nature methods 15, 53–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner RA, Siuzdak G, Prospero-Garcia O, Henriksen SJ, Boger DL, and Cravatt BF (1994). Cerebrodiene: a brain lipid isolated from sleep-deprived cats. Proceedings of the National Academy of Sciences 91, 9505–9508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schäberle TF, Hughes DE, Epstein S, et al. (2015). A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipmann F, and Kaplan NO (1946). A COMMON FACTOR IN THE ENZYMATIC ACETYLATION OF SULFANILAMIDE AND OF CHOLINE. Journal of Biological Chemistry 162, 743–744. [Google Scholar]
- Liu Y, Mrzic A, Meysman P, De Vijlder T, Romijn EP, Valkenborg D, Bittremieux W, and Laukens K (2020). MESSAR: Automated recommendation of metabolite substructures from tandem mass spectra. PloS one 15, e0226770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodge S, Nitschke P, Loo RL, Kimhofer T, Bong SH, Richards T, Begum S, Spraul M, Schaefer H, Lindon JC, et al. (2021). Low Volume in Vitro Diagnostic Proton NMR Spectroscopy of Human Blood Plasma for Lipoprotein and Metabolite Analysis: Application to SARS-CoV-2 Biomarkers. Journal of proteome research 20, 1415–1423. [DOI] [PubMed] [Google Scholar]
- Loos M, Gerber C, Corona F, Hollender J, and Singer H (2015). Accelerated isotope fine structure calculation using pruned transition trees. Analytical Chemistry 87, 5738–5744. [DOI] [PubMed] [Google Scholar]
- Loos M, and Singer H (2017). Nontargeted homologue series extraction from hyphenated high resolution mass spectrometry data. Journal of cheminformatics 9, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig C, and Günther UL (2011). MetaboLab--advanced NMR data processing and analysis for metabolomics. BMC bioinformatics 12, 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahieu NG, Huang X, Chen Y Jr., and Patti GJ (2014). Credentialing Features: A Platform to Benchmark and Optimize Untargeted Metabolomic Methods. Analytical Chemistry 86, 9583–9589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder EL, Billings EM, Benton HP, Martin RL, Palermo A, Guijas C, Rinschen MM, Domingo-Almenara X, Montenegro-Burke JR, Tagtow BA, et al. (2021). Cognitive analysis of metabolomics data for systems biology. Nat Protoc 16, 1376–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmström J, Lee H, and Aebersold R (2007). Advances in proteomic workflows for systems biology. Current opinion in biotechnology 18, 378–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchester KL (1998). Albert Szent-Györgyi and the unravelling of biological oxidation. Trends in Biochemical Sciences 23, 37–40. [DOI] [PubMed] [Google Scholar]
- Marquis E (1896). Über den Verbleib des Morphin im tierischen Organismus. In Pharmazeutische Zentralhalle für Deutschland (Jurjew, Arb.: Der Pharm. Inst. zu Dorpat), p. 117. [Google Scholar]
- Martin AJ, and Synge RL (1941). A new form of chromatogram employing two liquid phases: A theory of chromatography. 2. Application to the micro-determination of the higher monoamino-acids in proteins. The Biochemical journal 35, 1358–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachran AD, Balabin I, Cathey T, Transue TR, Al-Ghoul H, Grulke C, Sobus JR, and Williams AJ (2019). Linking in silico MS/MS spectra with chemistry data to improve identification of unknowns. Scientific Data 6, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra BB (2021). New software tools, databases, and resources in metabolomics: updates from 2020. Metabolomics 17, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulheirn G (2000). Robinson, Woodward and the synthesis of cholesterol. Endeavour 24, 107–110. [Google Scholar]
- Nagana Gowda GA, and Raftery D (2015). Can NMR solve some significant challenges in metabolomics? Journal of magnetic resonance (San Diego, Calif. : 1997) 260, 144–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagana Gowda GA, and Raftery D (2021). NMR-Based Metabolomics. Advances in experimental medicine and biology 1280, 19–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DH, Nguyen CH, and Mamitsuka H (2019). Recent advances and prospects of computational methods for metabolite identification: a review with emphasis on machine learning approaches. Briefings in bioinformatics 20, 2028–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, and Pettersson S (2012). Host-gut microbiota metabolic interactions. Science (New York, N.Y.) 336, 1262–1267. [DOI] [PubMed] [Google Scholar]
- Pfeuffer J, Sachsenberg T, Alka O, Walzer M, Fillbrunn A, Nilse L, Schilling O, Reinert K, and Kohlbacher O (2017). OpenMS - A platform for reproducible analysis of mass spectrometry data. Journal of biotechnology 261, 142–148. [DOI] [PubMed] [Google Scholar]
- Plumb RS, McDonald T, Rainville PD, Hill J, Gethings LA, Johnson KA, and Wilson ID (2021). High-Throughput UHPLC/MS/MS-Based Metabolic Profiling Using a Vacuum Jacketed Column. Analytical Chemistry. [DOI] [PubMed] [Google Scholar]
- Purcell EM, Torrey HC, and Pound RV (1946). Resonance Absorption by Nuclear Magnetic Moments in a Solid. Physical Review 69, 37–38. [Google Scholar]
- Rabi II, Zacharias JR, Millman S, and Kusch P (1938). A New Method of Measuring Nuclear Magnetic Moment. Physical Review 53, 318–318. [DOI] [PubMed] [Google Scholar]
- Reinitzer F (1888). Beiträge zur Kenntniss des Cholesterins. Monatshefte für Chemie und verwandte Teile anderer Wissenschaften 9, 421–441. [Google Scholar]
- Reinitzer F (1989). Contributions to the knowledge of cholesterol. Liquid Crystals 5, 7–18. [Google Scholar]
- Rinschen MM, Ivanisevic J, Giera M, and Siuzdak G (2019). Identification of bioactive metabolites using activity metabolomics. Nature reviews. Molecular cell biology 20, 353–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers MA, Villareal VA, Schaefer EA, Peng LF, Corey KE, Chung RT, and Yang PL (2012). Lipid metabolite profiling identifies desmosterol metabolism as a new antiviral target for hepatitis C virus. Journal of the American Chemical Society 134, 6896–6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röntgen WC (1898). Ueber eine neue Art von Strahlen. Annalen der Physik 300, 12–17. [Google Scholar]
- Röst HL, Sachsenberg T, Aiche S, Bielow C, Weisser H, Aicheler F, Andreotti S, Ehrlich HC, Gutenbrunner P, Kenar E, et al. (2016). OpenMS: a flexible open-source software platform for mass spectrometry data analysis. Nature methods 13, 741–748. [DOI] [PubMed] [Google Scholar]
- Rutherford E, and Geiger H (1908). An electrical method of counting the number of α-particles from radioactive substances. Proceedings of the Royal Society of London. Series A, Containing Papers of a Mathematical and Physical Character 81, 141–161. [Google Scholar]
- Salek RM, Steinbeck C, Viant MR, Goodacre R, and Dunn WB (2013). The role of reporting standards for metabolite annotation and identification in metabolomic studies. GigaScience 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scapin G, Potter CS, and Carragher B (2018). Cryo-EM for Small Molecules Discovery, Design, Understanding, and Application. Cell Chemical Biology 25, 1318–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlienger JL (2012). L’édifiante histoire du cholestérol : de la pierre de fiel au récepteur aux LDL: The edifying cholesterol story: From ‘gall stone’ to the LDL receptor. Médecine des Maladies Métaboliques 6, 97–103. [Google Scholar]
- Schoenheimer R, and Rittenberg D (1935). DEUTERIUM AS AN INDICATOR IN THE STUDY OF INTERMEDIARY METABOLISM. I. Journal of Biological Chemistry 111, 163–168. [DOI] [PubMed] [Google Scholar]
- Shin S-Y, Fauman EB, Petersen A-K, Krumsiek J, Santos R, Huang J, Arnold M, Erte I, Forgetta V, Yang T-P, et al. (2014). An atlas of genetic influences on human blood metabolites. Nature Genetics 46, 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine EE, Xue M, Patel JR, Healy AR, Surovtseva YV, Herzon SB, and Crawford JM (2018). Model Colibactins Exhibit Human Cell Genotoxicity in the Absence of Host Bacteria. ACS chemical biology 13, 3286–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov A, Qiu Y, Jia W, Walker DI, Jones DP, and Du X (2019). ADAP-GC 4.0: Application of Clustering-Assisted Multivariate Curve Resolution to Spectral Deconvolution of Gas Chromatography–Mass Spectrometry Metabolomics Data. Analytical Chemistry 91, 9069–9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, and Siuzdak G (2005). METLIN: a metabolite mass spectral database. Therapeutic drug monitoring 27, 747–751. [DOI] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O'Maille G, Abagyan R, and Siuzdak G (2006). XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Analytical Chemistry 78, 779–787. [DOI] [PubMed] [Google Scholar]
- Spann NJ, Garmire LX, McDonald JG, Myers DS, Milne SB, Shibata N, Reichart D, Fox JN, Shaked I, Heudobler D, et al. (2012). Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell 151, 138–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, Merrill AH Jr., Murphy RC, Raetz CR, Russell DW, et al. (2007). LMSD: LIPID MAPS structure database. Nucleic Acids Res 35, D527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teranishi R, Mon TR, Robinson AB, Cary P, and Pauling L (1972). Gas chromatography of volatiles from breath and urine. Analytical Chemistry 44, 18–20. [DOI] [PubMed] [Google Scholar]
- Thaulow MCJ (1838). Ueber die Zuckersäure. Annalen der Pharmacie 27, 113–130. [Google Scholar]
- Thomas MC, Mitchell TW, Harman DG, Deeley JM, Nealon JR, and Blanksby SJ (2008). Ozone-Induced Dissociation: Elucidation of Double Bond Position within Mass-Selected Lipid Ions. Analytical Chemistry 80, 303–311. [DOI] [PubMed] [Google Scholar]
- Tollens B (1882). Ueber ammon-alkalische Silberlösung als Reagens auf Aldehyd. Berichte der deutschen chemischen Gesellschaft 15, 1635–1639. [Google Scholar]
- Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, Kanazawa M, VanderGheynst J, Fiehn O, and Arita M (2015). MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nature methods 12, 523–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugawa H, Ikeda K, Takahashi M, Satoh A, Mori Y, Uchino H, Okahashi N, Yamada Y, Tada I, Bonini P, et al. (2020). A lipidome atlas in MS-DIAL 4. Nature biotechnology 38, 1159–1163. [DOI] [PubMed] [Google Scholar]
- van de Velde B, Guillarme D, and Kohler I (2020). Supercritical fluid chromatography - Mass spectrometry in metabolomics: Past, present, and future perspectives. Journal of chromatography. B, Analytical technologies in the biomedical and life sciences 1161, 122444. [DOI] [PubMed] [Google Scholar]
- van der Hooft JJJ, Wandy J, Barrett MP, Burgess KEV, and Rogers S (2016). Topic modeling for untargeted substructure exploration in metabolomics. Proceedings of the National Academy of Sciences 113, 13738–13743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven A, Giera M, and Mayboroda OA (2020). Scientific workflow managers in metabolomics: an overview. The Analyst 145, 3801–3808. [DOI] [PubMed] [Google Scholar]
- Vizcaino MI, and Crawford JM (2015). The colibactin warhead crosslinks DNA. Nature Chemistry 7, 411–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino MI, Engel P, Trautman E, and Crawford JM (2014). Comparative Metabolomics and Structural Characterizations Illuminate Colibactin Pathway-Dependent Small Molecules. Journal of the American Chemical Society 136, 9244–9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T, et al. (2016). Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nature biotechnology 34, 828–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warth B, Spangler S, Fang M, Johnson CH, Forsberg EM, Granados A, Martin RL, Domingo-Almenara X, Huan T, Rinehart D, et al. (2017). Exposome-Scale Investigations Guided by Global Metabolomics, Pathway Analysis, and Cognitive Computing. Analytical Chemistry 89, 11505–11513. [DOI] [PubMed] [Google Scholar]
- Watts G (2015). Carl Djerassi. The Lancet 385, 600. [DOI] [PubMed] [Google Scholar]
- Williams RJ (1956). Biochemical Individuality. The Basis for the Genetotrophic Concept. (New York: John Willey & Sons; ). [Google Scholar]
- Wilson MR, Jiang Y, Villalta PW, Stornetta A, Boudreau PD, Carrá A, Brennan CA, Chun E, Ngo L, Samson LD, et al. (2019). The human gut bacterial genotoxin colibactin alkylates DNA. Science (New York, N.Y.) 363, eaar7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windaus A (1932). Über die Konstitution des Cholesterins und der Gallensäuren. Hoppe-Seyler's Zeitschrift Fur Physiologische Chemie 213, 147–187. [Google Scholar]
- Wishart DS (2016). Emerging applications of metabolomics in drug discovery and precision medicine. Nature reviews. Drug discovery 15, 473–484. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, Sajed T, Johnson D, Li C, Karu N, et al. (2018). HMDB 4.0: the human metabolome database for 2018. Nucleic acids research 46, D608–d617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods KJP, and McDermott JH (2018). Schema learning for the cocktail party problem. Proceedings of the National Academy of Sciences 115, E3313–E3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward RB, Sondheimer F, and Taub D (1951). THE TOTAL SYNTHESIS OF CHOLESTEROL. Journal of the American Chemical Society 73, 3548–3548. [Google Scholar]
- Woolley DW (1959). Antimetabolites. They help in discovery of metabolic pathways and in the understanding and treatment of some diseases 129, 615–621. [DOI] [PubMed] [Google Scholar]
- Xie C, Gu L, Wu Q, Li L, Wang C, Yu J, and Tang K (2021). Effective Chiral Discrimination of Amino Acids through Oligosaccharide Incorporation by Trapped Ion Mobility Spectrometry. Analytical Chemistry 93, 859–867. [DOI] [PubMed] [Google Scholar]
- Xue J, Guijas C, Benton HP, Warth B, and Siuzdak G (2020). METLIN MS(2) molecular standards database: a broad chemical and biological resource. Nature methods 17, 953–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M, Kim CS, Healy AR, Wernke KM, Wang Z, Frischling MC, Shine EE, Wang W, Herzon SB, and Crawford JM (2019). Structure elucidation of colibactin and its DNA cross-links. Science (New York, N.Y.) 365, eaax2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Soga T, and Pollard PJ (2013). Oncometabolites: linking altered metabolism with cancer. The Journal of clinical investigation 123, 3652–3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yore MM, Syed I, Moraes-Vieira PM, Zhang T, Herman MA, Homan EA, Patel RT, Lee J, Chen S, Peroni OD, et al. (2014). Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell 159, 318–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni N, Saghatelian A, and Patti GJ (2015). Defining the metabolome: size, flux, and regulation. Molecular cell 58, 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Desai A, Yang SY, Bae KB, Antczak MI, Fink SP, Tiwari S, Willis JE, Williams NS, Dawson DM, et al. (2015). TISSUE REGENERATION. Inhibition of the prostaglandin-degrading enzyme 15-PGDH potentiates tissue regeneration. Science (New York, N.Y.) 348, aaa2340. [DOI] [PMC free article] [PubMed] [Google Scholar]