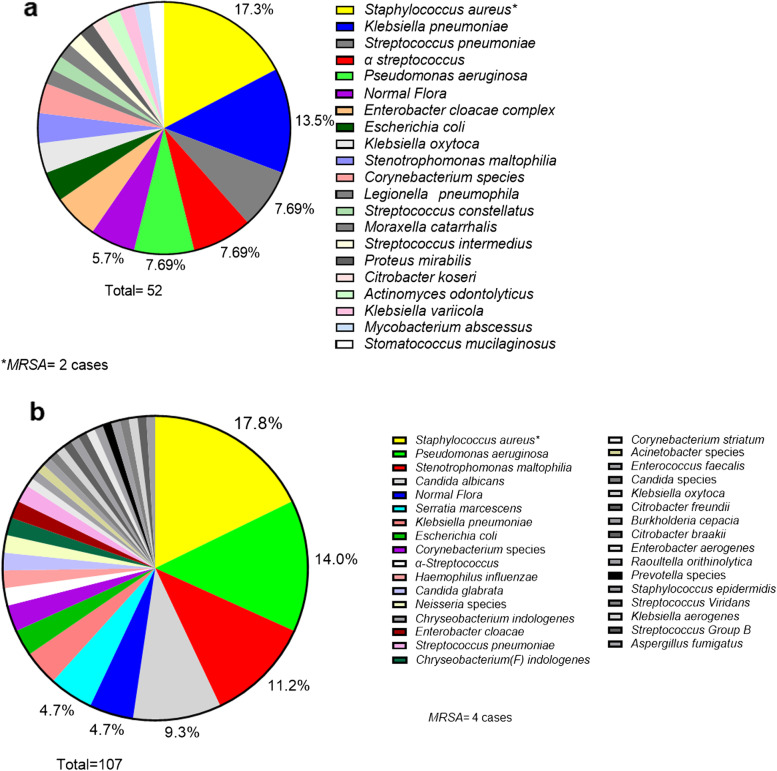
Fig. 2.
Bacterial pathogens identified in respiratory bacterial co-infection and secondary infection cases with coronavirus disease 2019. a All identified organisms as a proportion of total number of organisms per pathogen in sputum culture and urinary antigen of co-infection pneumonia with coronavirus disease 2019 (COVID-19). Bacterial pathogens detected in COVID-19 patients with respiratory bacterial co-infections, as a proportion (%) of the total number of isolates (n = 47). Some patients had multiple bacterial infection. In 24 cases, no causative organism was detected in sputum cultures. MRSA, Methicillin-Resistant Staphylococcus aureus. b All identified organisms as a proportion of total number of organisms per pathogen in sputum, tracheal aspirate, or bronchoalveolar lavage culture of secondary infection pneumonia with COVID-19. Bacterial pathogens detected in COVID-19 patients with respiratory bacterial secondary infection, as a proportion (%) of the total number of isolates (n = 107). Some patients had more than one bacterial infection. MRSA, Methicillin-Resistant Staphylococcus aureus