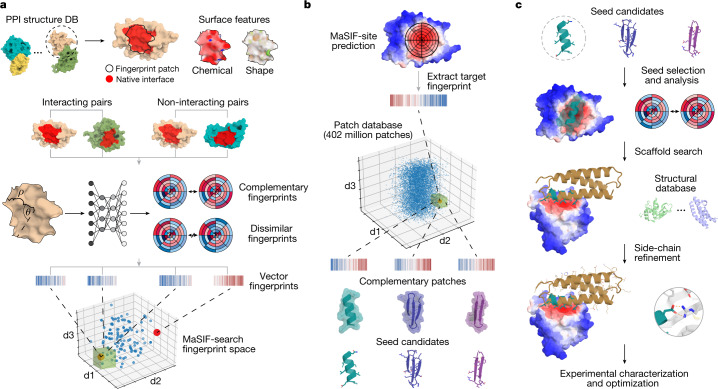
Fig. 1. Surface-centric design of de novo site-specific protein binders.
a, Schematic of fingerprint generation. Protein binding sites are spatially embedded as vector fingerprints. Protein surfaces are decomposed into overlapping radial patches, and a neural network trained on native interacting protein pairs learns to embed the fingerprints such that complementary fingerprints are placed in a similar region of space. We show an illustration for a subsample of the fingerprints projected in a space reduced to three dimensions. The green box highlights a region of complementary fingerprints. b, MaSIF-seed—a method to identify new binding seeds. A target patch is identified by MaSIF-site based on the propensity to form buried interfaces. Using MaSIF-seed, fingerprint complementarity is evaluated between the target patch and all fingerprints in a large database (around 402 million patches); the pairs of fingerprints are subsequently ranked. The top patches are aligned and rescored to enable a more precise evaluation of the seed candidates. c, Scaffold search, seed grafting and interface redesign. The selected seeds are transferred to protein scaffolds and the rest of the interface is redesigned using Rosetta. The top designs are selected and tested experimentally.