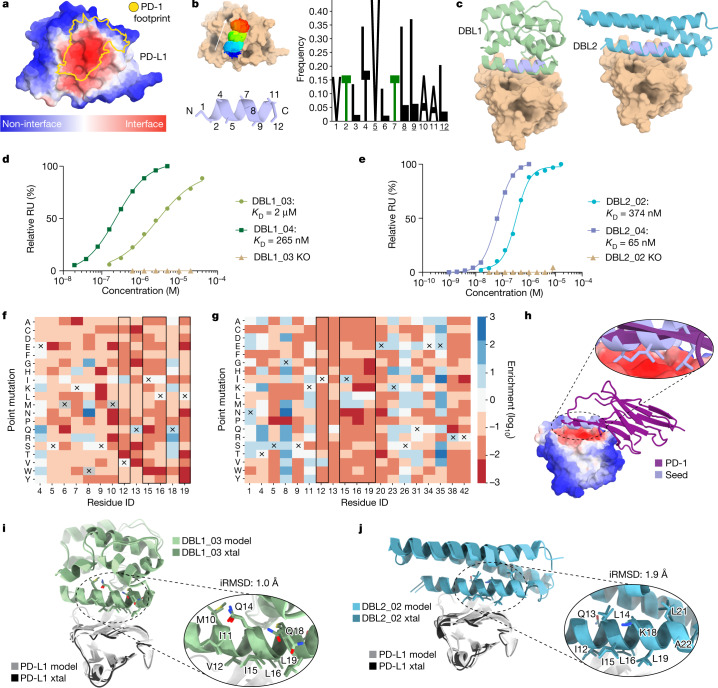
Fig. 3. De novo design and optimization of PD-L1 binders targeting a flat surface.
a, MaSIF-site prediction of the interface propensity of PD-L1. The predicted interface (red) overlaps with the binding site of the native interaction partner PD-1 (yellow). b, Helical seeds were predicted by MaSIF-seed and clustered. The dominant cluster showed strong amino acid preferences (Z-score > 2). Hotspot residues are underlined. c, Binders based on two different scaffold proteins using the selected seed were identified. d, The binding affinities of DBL1 designs after combinatorial (light green) and SSM library optimization (dark green), measured using SPR. Mutation of a hotspot residue (V12R) ablates binding of DBL1_03 (wheat). e, The binding affinities of DBL2 designs after combinatorial (light blue) and SSM library optimization (dark blue), measured using SPR. Mutation of a hotspot residue (V12R) knocks out binding of DBL2_02 (wheat). f, SSM analysis of regions of interest in the binding interface of DBL1_03. The original residue of DBL1_03 is indicated by a cross and hotspot residue positions are shown in black boxes. Enrichment in the binding population (blue) and in the non-binding population (red) is indicated. g, SSM data in the binding interface of DBL2_03. The original residue of DBL2_02 is indicated by a cross. h, The binding mode of the selected seed in comparison to the native interaction partner PD-1. i, Crystal (xtal) structure of DBL1_03 in a complex with PD-L1. The computational model (light green) is aligned with the crystal structure (dark green). Inset: the alignment of the residues in the binding seed. j, Crystal structure of DBL2_02 in a complex with PD-L1, shown by aligning the computational model (light blue) with the crystal structure (dark blue). Inset: the alignment of the residues in the binding seed represented as sticks.