Figure 5.
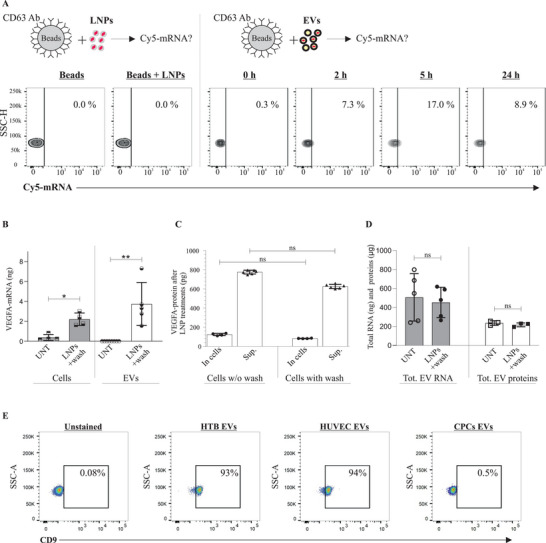
Characterization of extracellular vesicles (EVs) and validation of LNP‐mRNA Incorporation into EVs. A) Flow cytometry analysis showing the percentage of purified CD63+ EVs containing Cy5‐eGFP mRNA isolated from LNP‐treated cells, at different time points (n = 2). One representative measurement is shown from each time point. LNP‐Cy5‐eGFP mRNA + beads were used instead of EVs+beads, to evaluate whether a contamination had occurred. For the negative control, only beads (without EVs or LNPs) were used. B) Detection of LNP‐VEGF‐A mRNA in cells and their secreted EVs, after washing out the LNPs (n = 6 for LNP‐treated and UNT (untreated) cells, n = 5 for treated EVs and n = 8 for UNT EVs). Statistically significant differences were evaluated by the Mann‐Whitney U‐test (*p < 0.05; **p < 0.01). C) Detection of VEGF‐A protein in cell lysates and supernatants, after washing out LNPs. The Kruskal‐Wallis test followed by Dunn's multiple comparison test was used to compare washed versus unwashed supernatants and washed versus unwashed cells (ns = no significant differences). D) The amounts of total EV‐RNA and total EV‐proteins between washed and unwashed cells from LNP‐treated or untreated groups (n = 6). Statistically significant differences were evaluated by the Mann‐Whitney U‐test (ns = no significant differences). E) Detection of CD63, and CD9‐positive HTB‐, HUVEC‐, and CPC‐EVs. Pre‐enriched EVs were captured by anti‐CD63‐antibody‐conjugated beads, and then these CD63+ EVs were captured/isolated with PE‐CD9 antibody and acquired on a BD FACSLyric system (BD Biosciences). Results show that EVs that were CD63‐positive were also CD9‐positive. Data were analyzed using FlowJo software (TreeStar Inc.). As a negative control, CD63‐antibody‐conjugated beads alone (without EVs) were incubated with an equivalent volume of PBS. The experiment was performed in biological duplicates (n = 2). One representative measurement from each EV type is shown.
