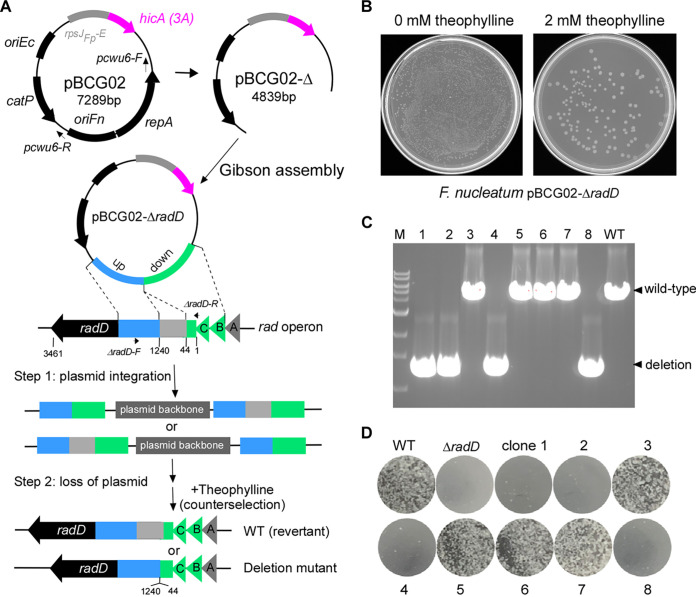
FIG 3.
Applying hicA(3A) as a counterselectable marker for gene deletion in F. nucleatum. (A) Graphic representation of producing the nonreplicative vector backbone pBCG02-Δ used to generate unmarked in-frame gene deletions in F. nucleatum via a two-step strategy. The primer pair (pCWU6-F/pCWU6-R, indicated by short arrows) was used to perform inverse PCR to remove the region, including the oriFn and repA. The radABCD operon is shown. Approximately 1.6-kb fragments homologous to the upstream (blue) and downstream (green) regions of the designed deletion region in radD were amplified by PCR. They have been ligated adjacently on the vector backbone pBCG02-Δ via the Gibson assembly procedure. The resulting plasmid, pBCG02-ΔradD, was used to generate an unmarked radD deletion (the gene deletion region coding for the amino acids in positions between 44 and 1240 of the RadD protein of 3,461 amino acids) mutant in the wild-type background by a two-step allelic exchange. (B) TSPC agar plate representative of the counterselection effect by putative HicA(3A) toxin expression in F. nucleatum. A 100-μL aliquot of 1,000-fold-diluted overnight culture of the pBCG02-pBCG02-ΔradD-integrated strain was plated on TSPC agar plates containing 0 or 2 mM inducer theophylline to select for cells in which the plasmid had been excised and lost. (C) Screen of radD deletion mutants by colony PCR with the primer pair ΔradD-F/ΔradD-R (indicated in panel A). Lanes 1 to 8, 10 μL of PCR products of eight colonies randomly selected from the TSPC agar plate. Four of the eight chosen random colonies were deletion mutants, while four reverted to wild type during the second recombination event. Bands of approximately 1.4 kb indicate radD deletion, whereas bands of about 5.0 kb indicate a wild-type (WT) genotype. (D) F. nucleatum wild-type, radD mutant strain, and 8 counterselection isolates from panel C were examined for coaggregation with A. oris MG-1. A representative result was presented after these experiments were carried out three times.