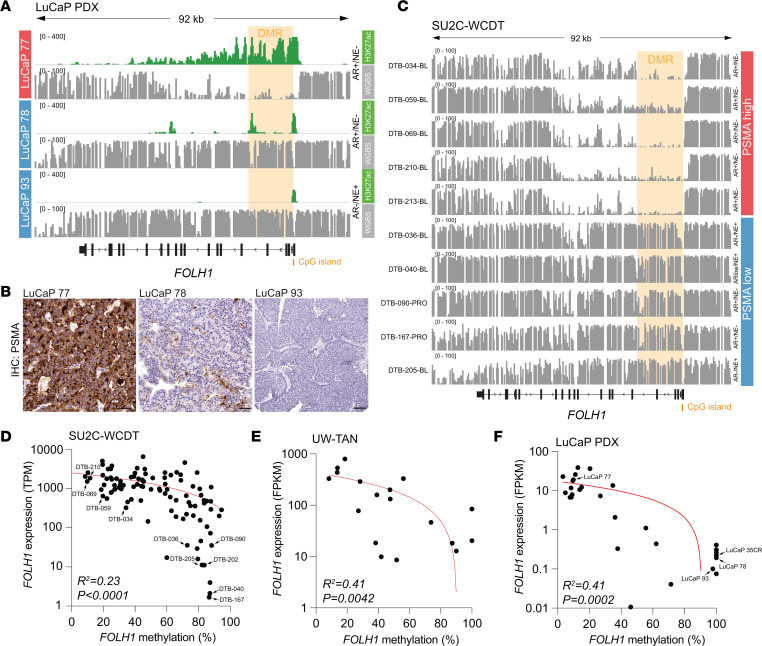
Figure 6. Epigenetic changes enforce silencing of FOLH1/PSMA.
(A) Whole-genome bisulfite sequencing (WGBS) tracks from PSMA-high (LuCaP 77, red) and PSMA-negative (LuCaP 78 and 93, blue) tumors reveal a differentially methylated region (DMR) encompassing the first 14 kb of the FOLH1 gene and inverse enrichment for H3K27ac. (B) Micrographs of PSMA IHC of LuCaP 77, LuCaP 78, and LuCaP 93 demonstrate the difference in PSMA expression. (C) Representative WGBS tracks of mCRPC tumors from the SU2C-WCDT cohort show gain of methylation in the DMR in PSMA-low/negative tumors. (D–F) Scatter plots show the correlation between FOLH1 expression (based on RNA-Seq) and DMR methylation derived from WGBS (SU2C-WCDT, n = 98) (D) and targeted COMPARE-MS analyses UW-TAN (n = 18) (E) and LuCaP (n = 29) (F) cohorts. Curves were fit by linear regression, and R2 and P values were derived by Pearson correlation.