Figure 1.
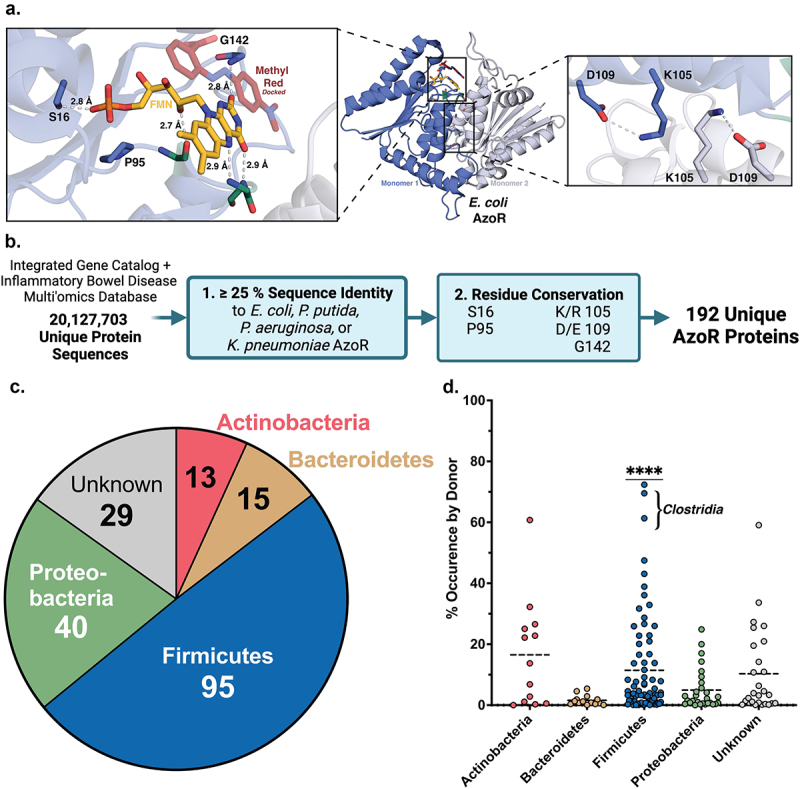
Structural metagenomics pipeline used to identify gut bacterial AzoR Enzymes. (a) Conserved residues (blue) used to define two-step rubric to identify AzoR proteins shown in EcAzoR (PDB: 2D5I), showing placement of FMN (yellow) and methyl red (red) docked as an example ligand. (b) Workflow of structure-guided rubric used to identify AzoR proteins across the IGC and IBDMDB. (c) Phylogenetic distribution of unique putative AzoR proteins across IGC and IBDMDB. (d) Occurrence for each AzoR sequence per Individual across IGC arranged by phyla, ****P < 0.0001 by a one sample t test comparing Firmicutes to all other groups.
