Figure 3.
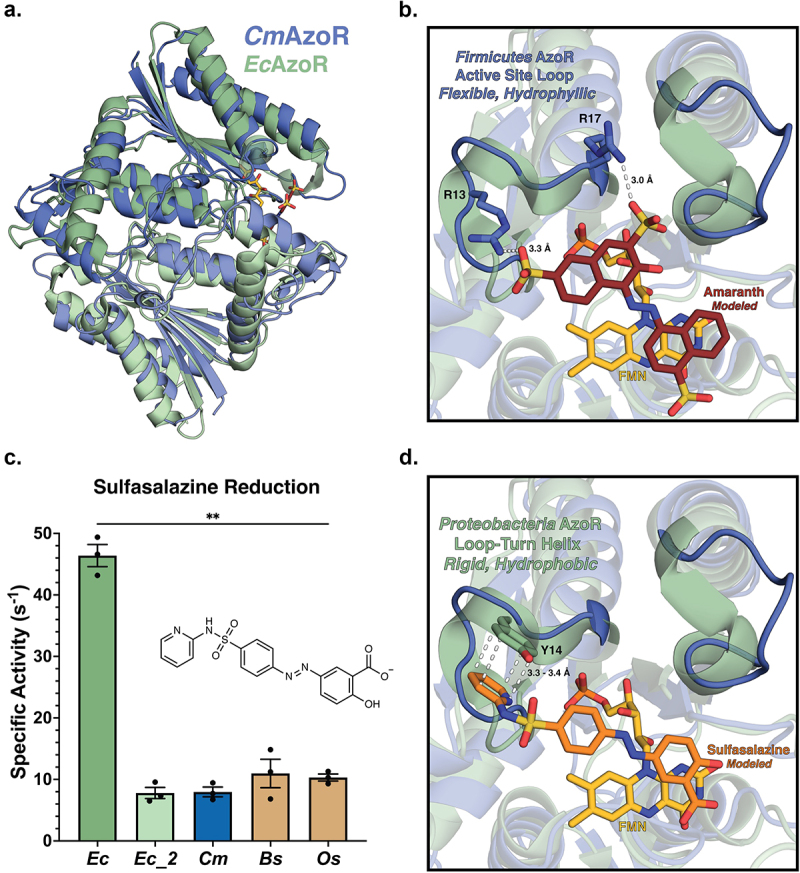
AzoR active site environments point to substrate specificity. (a) Homodimers of crystal structures of EcAzoR (green) and CmAzoR (blue) overlaid and analysis of active site structural disparities. FMN (yellow) is located according to the structure of EcAzoR (PDB: 2D5I). Amaranth (red) was modeled into the active site using methyl red in the crystal structure of PaAzoR (PDB: 2V9C) as a reference for ligand placement. (b) Active site docked models for Amaranth (B) and Sulfasalazine (d) azo reduction by purified AzoR enzymes. Ligands were docked into active sites of crystal structures to point toward structural differences driving differential enzyme activity. Error bars represent SEM across three biological replicates. Catalytic efficiencies and active site docked models for Sulfasalazine (c) azo reduction by purified AzoR enzymes. Statistical comparisons reflect activity of CmAzoR compared to other enzymes and were calculated using Tukey’s multiple comparisons test, **P < 0.01.
