Figure 4.
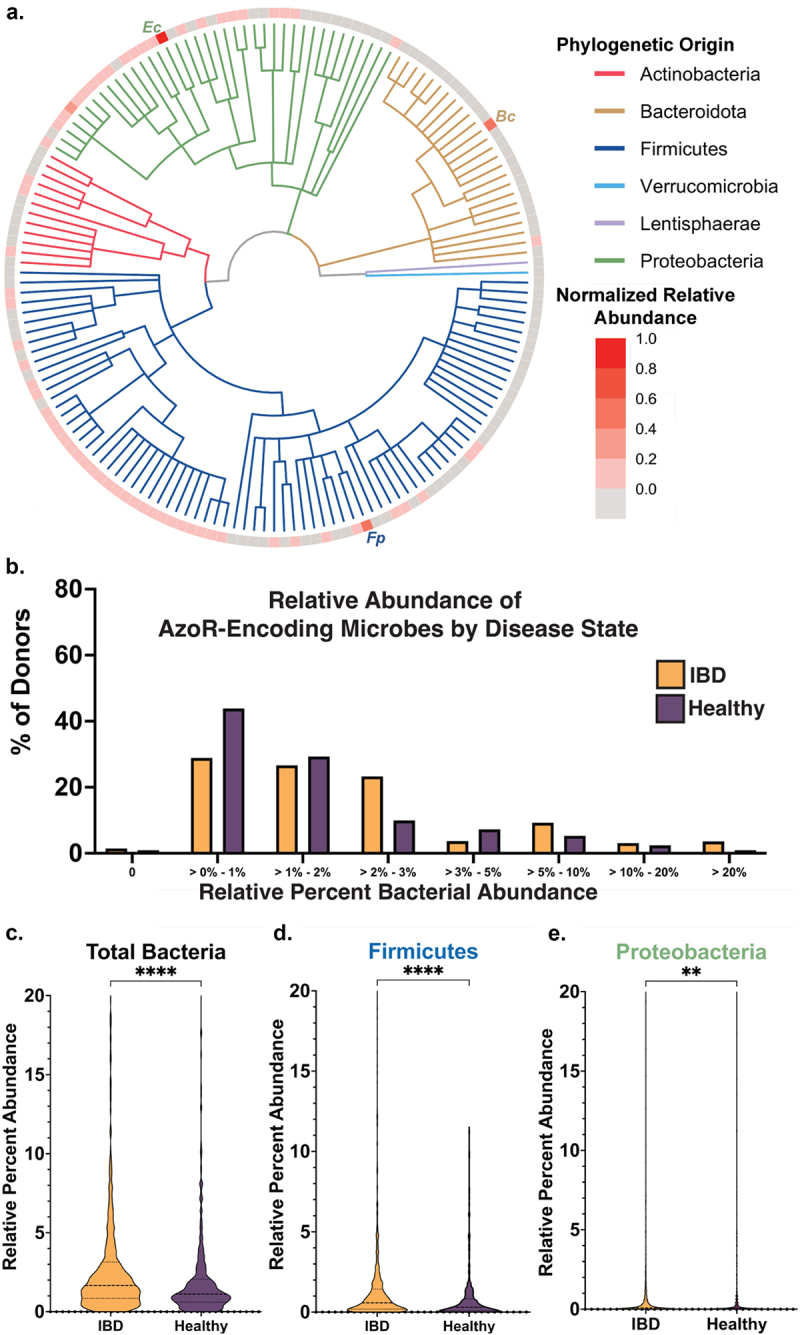
Azo—encoding microbes are sparse in health and disease. (a) Cladogram reflecting microbes used for integrative analysis query, with abundances for each relative to other AzoR-encoding strains. Percent of percent of donors containing AzoR-encoding microbes as determined by Integrated Search space combining EC Annotations, extant AzoR-encoding microbes reported in literature, and species identified using structural metagenomics in this study for total bacterial abundance. (b) Abundance of Percent of percent of donors containing AzoR-producing microbes at a range of relative abundance levels across the x-axis, queried using an Integrated Search space combining EC Annotations, extant AzoR-producing microbes reported in literature, and species identified using structural metagenomics in this study for total bacterial abundance. Violin plots comparing AzoR abundances between IBD patients and healthy individuals across the IBDMDB, comparing relative % abundances for all microbes (c), Firmicutes (d), and Proteobacteria (e) that produce AzoR genes as annotated by Integrative Analysis used in this manuscript. ****P < 0.0001, **P < 0.01 by an unpaired t test.
