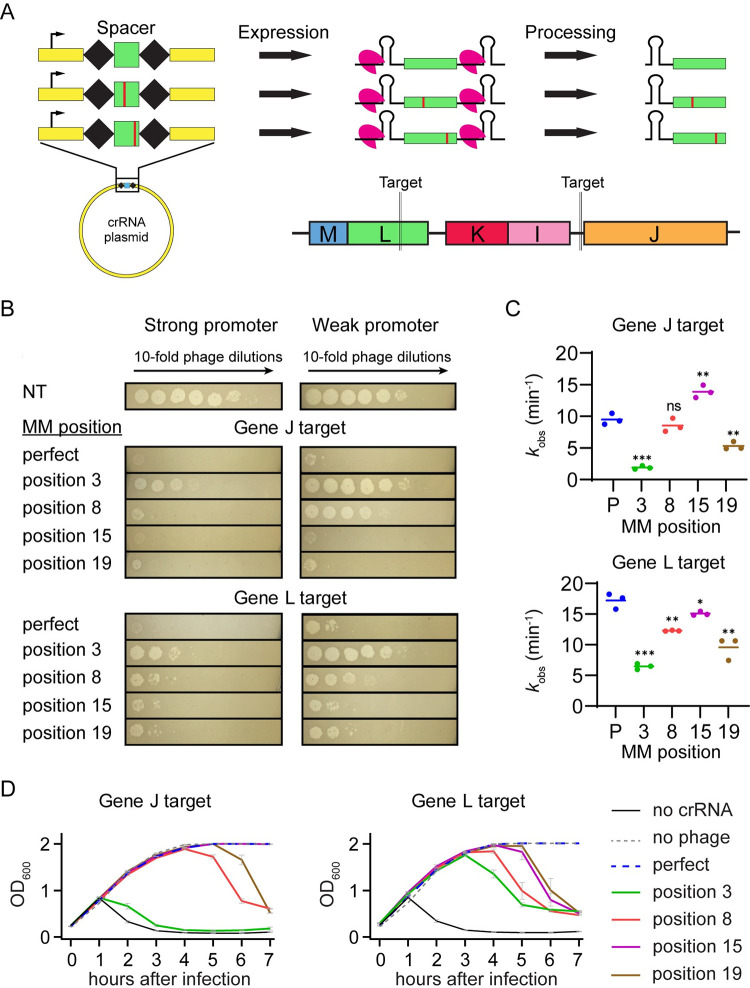
Fig 1. Effects of mismatched crRNAs on Cas12a-mediated phage defense.
(A) Schematic of crRNA expression and processing by FnCas12a and crRNA phage target locations. crRNA mismatches were introduced by mutating individual nucleotides in the spacer sequence. After expression of the pre-crRNA, Cas12a processes it into a guiding crRNA that partially matches the lambda phage genome targets upstream of gene J and in the coding region of gene L. See S1a for target and crRNA spacer sequences. (B) Measurement of phage protection provided by crRNAs with and without target mismatches. Spot assays were performed with bacteria expressing FnCas12a and a crRNA construct that either perfectly matches the lambda phage genome (perfect) or has a crRNA mismatch (MM) at a position in the spacer (position x, sequences shown in S1A Fig). A non-targeting crRNA construct (NT) was used as a negative control. Lambda phage was spotted on cells with 10-fold decreasing concentration at each spot going from left to right. Expression of FnCas12a and pre-crRNAs were controlled by a stronger inducible PBAD promoter or a weaker constitutive promoter. (C) Observed rate constants for in vitro cleavage by Cas12a armed with crRNAs containing target mismatches. Plasmids bearing target sequences for gene J or L were used to measure Cas12a cleavage. Mismatch positions or perfect crRNAs (P) are indicated on the horizontal axis. Data from 3 replicates are plotted. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, ns = no significant difference compared to the perfect crRNA based on unpaired two-tailed t test. See S1B and S1C Fig and S1 Data for gels and quantification. See S1 Fig for crRNA and target sequences, representative gels, and fit data. (D) Growth curves for E. coli expressing mismatched crRNAs following phage infection. Bacteria containing the PBAD FnCas12a expression plasmid and various crRNA expression plasmids were inoculated in liquid culture and induced immediately. Lambda phage was added 1.5 h after inoculation and OD600 measurements were taken every hour. Bacteria expressed no cRNA, a crRNA with no mismatches to the target (perfect) or a crRNA with a mismatch at the indicated position (position x). A no phage condition was performed as a negative control. The average of 2 replicates is plotted, with error bars representing standard deviation. See S2 Data for quantified data.