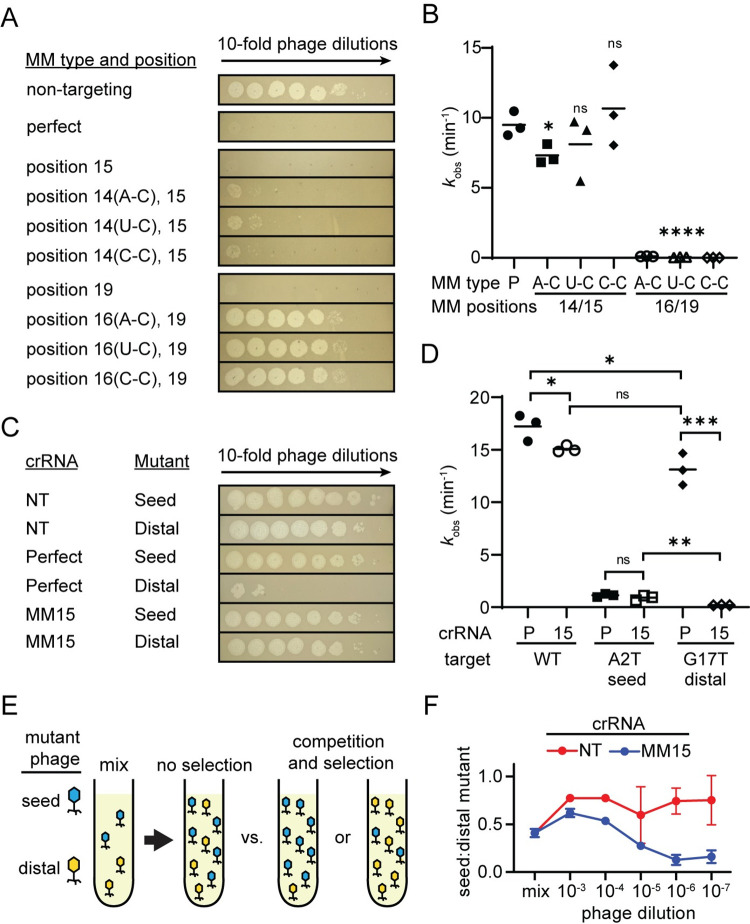
Fig 3. Two PAM-distal mismatches are more deleterious than seed mismatches.
(A) Spot assays performed using E. coli expressing FnCas12a and a crRNA that perfectly matches the lambda phage gene J target (perfect) or has mismatches at the indicated positions. Three types of second mismatches were added and the type of the mismatch is indicated in parenthesis next to the position number. See S7A Fig for crRNA spacer sequences. (B) Observed rate constants for in vitro cleavage by Cas12a armed with crRNAs containing 2 target mismatches. Cleavage was measured for plasmid DNA containing a gene J target. The types of mismatches for the second mismatch are indicated. * P ≤ 0.05, **** P ≤ 0.0001, ns P > 0.05 compared to the perfectly matching crRNA based on unpaired two-tailed t test. See S7B and S7C Fig and S1 Data for gels, and quantified and fit data. (C) Phage spot assays for target mutant phages isolated upon challenge with Cas12a programmed with a position 15 mismatched crRNA targeting gene L. Spot assays were performed with E. coli expressing a non-targeting crRNA (NT), a crRNA that perfectly matched wild-type phage (Perfect), or the crRNA with a mismatch at position 15 (MM15). Phage mutations were in the seed (A2T) or PAM-distal (G17T) region. (D) Observed rate constants for in vitro Cas12a cleavage of plasmids bearing wild-type (WT), seed mutant (A2T), or PAM-distal mutant (G17T) gene L target sequences. Cas12a cleavage was measured for both the perfectly matched crRNA (P) or the MM15 crRNA (15). Significance was tested pairwise for all crRNA/target combination by unpaired two-tailed t test. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, ns P > 0.05. Pairwise comparisons for which P value are not indicated had a P < 0.0001. See S8 Fig and S1 Data for crRNA and target sequences, gels, and quantified and fit data. (E) Schematic of competition assay. Two mutant phages, A2T and G17T, were mixed at approximately equal titers. E. coli expressing Cas12a and the position 15 mismatched crRNA were infected with a dilution series of the mutant phage mix. Lysates were harvested and the proportion of each mutant phage was determined by high-throughput sequencing. (F) Ratio of seed mutant (A2T) to PAM-distal mutant (G17T) following lysis of cultures infected with a dilution series of the mixed phage. E. coli expressed Cas12a and either a non-targeting (NT, red) or position 15 mismatched (MM15, blue) crRNA. “Mix” indicates the initial mixture of phage. The average of 3 replicates is shown, with error bars indicating standard deviation. See S6 Data for raw and quantified data.