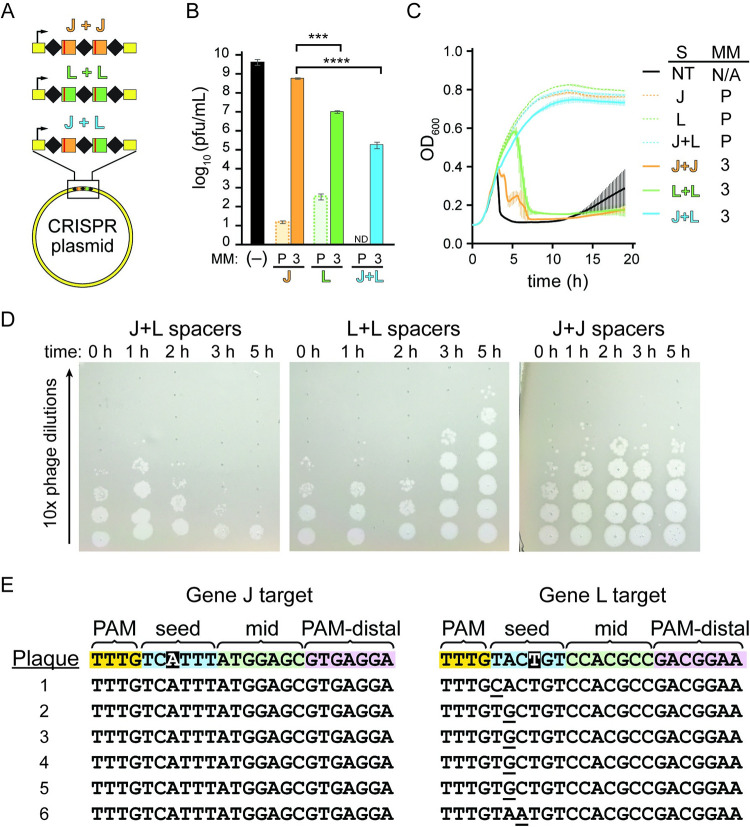
Fig 6. Multiple mismatched crRNAs provide more protection than individual mismatched crRNAs.
(A) Schematic of experiment in which 2 crRNAs bearing mismatches at position 3 are expressed from a CRISPR plasmid. The CRISPRs either have 2 identical spacers targeting gene J (J+J) or gene L (L+L), or have 2 different spacers targeting gene J and L (J+L). (B) Number of plaques formed on lawns of bacteria expressing multiple mismatched crRNAs. Plaque assays were performed using bacteria containing a plasmid expressing FnCas12a along with different crRNA expression plasmids. Plasmid expressed either the perfect crRNA (P) or the position 3 mismatched crRNA (3). For the perfect crRNAs, plasmids expressed either 1 (gene J or gene L alone) or 2 (J+L) spacers. For the position 3 mismatched crRNAs, each crRNA expression plasmid contains 2 spacers: both targeting gene J both targeting gene L and 1 spacer targeting each of gene J and gene L (J+L). (-) indicates a negative control in which no crRNA was expressed. Plaque forming units (pfu) was calculated using the number of plaques on each plate and the volume of phage lysate added. The average of multiple replicates (n = 2 to 4) is plotted with error bars representing standard deviation. Unpaired, two-tailed t tests were used to determine the statistical significance of each of the single spacer constructs compared to the double spacer construct, *** P ≤ 0.001, **** P ≤ 0.0001. See S2 Data for quantified data. (C) Growth curves using the same bacterial strains described in (A). Phage was added when the cells reached mid log phase at approximately 2 h after inoculation. The average of 3 replicates is plotted for each condition, with error bars representing standard deviation. See S2 Data for quantified data. (D) Spot assays used to measure the titer of phage over time in phage infection cultures. Spot assays were performed using 10-fold serial dilutions of phage harvested from cultures in (B) that infected bacterial strains with 2 mismatched spacers at different time points on lawns of CRISPR-inactive E. coli. (E) Sequences of both CRISPR targets in single phage plaques for phage harvested from E. coli cultures expressing a double spacer construct. The 2 crRNAs contained mismatches at positions highlighted in black. Target regions for the gene J and gene L target were sequenced for 6 individual plaques using Sanger sequencing. Target sequences are aligned to the WT sequence (top row) and mutations are underlined. See S12B Fig for chromatograms.