Figure 1.
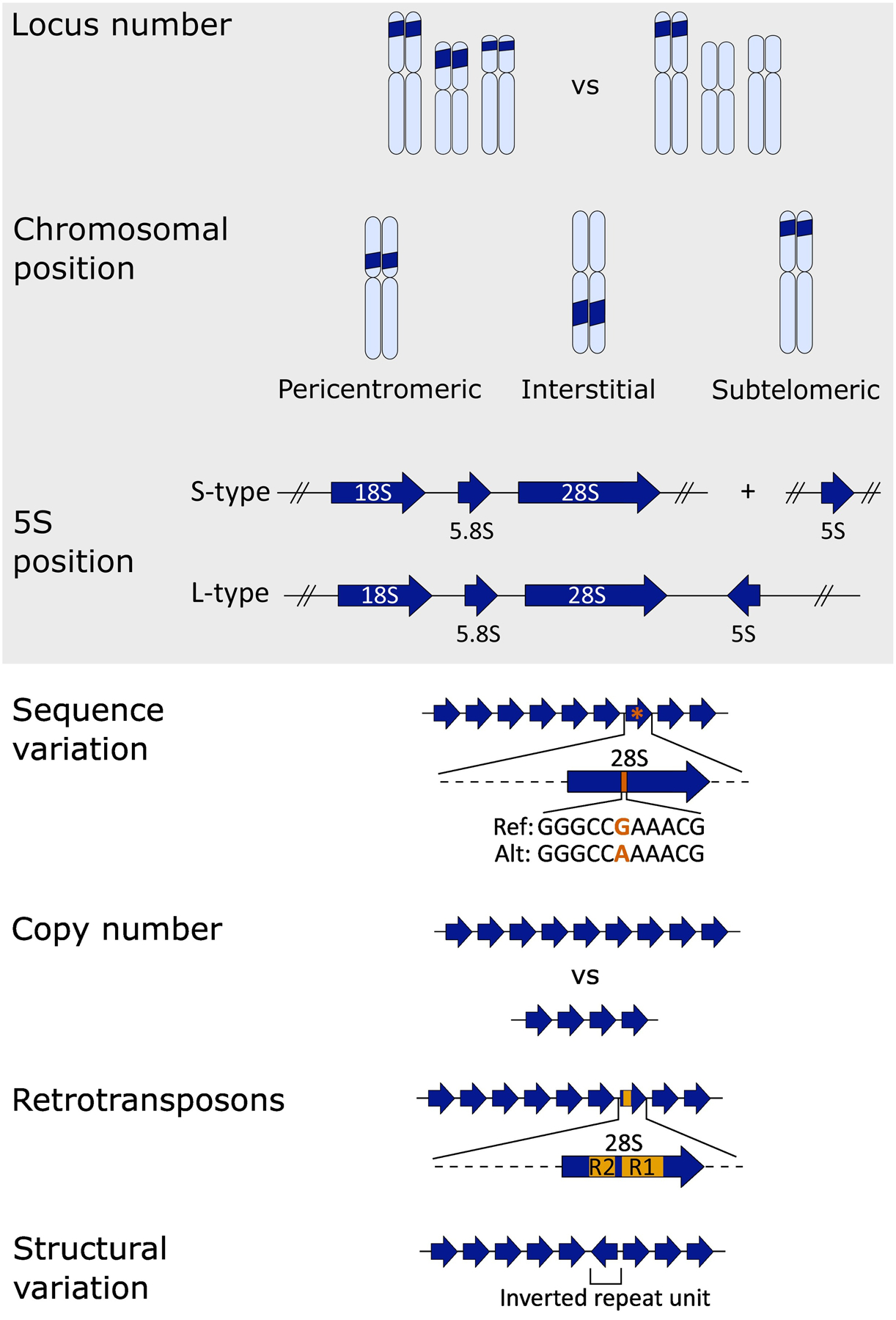
Some variation, such as the number of rDNA loci, chromosomal position of rDNA loci, and the position of the 5S with respect to the other rRNAs, have been readily ascertained for decades through use of in situ hybridization methods. Long-read sequencing has the potential to demystify rDNA variation types such as sequence variation, copy number variation, and structural variation – especially with regard to where different variants fall in the context of each rDNA array in the genome. In organisms with many rDNA arrays per genome (e.g., humans) long-read sequencing holds the potential to resolve the rDNA copy numbers of each array as well as the overall rDNA content per haploid genome, through ultralong reads that span whole arrays or by tiling long reads across the array using unique variants for anchoring.
