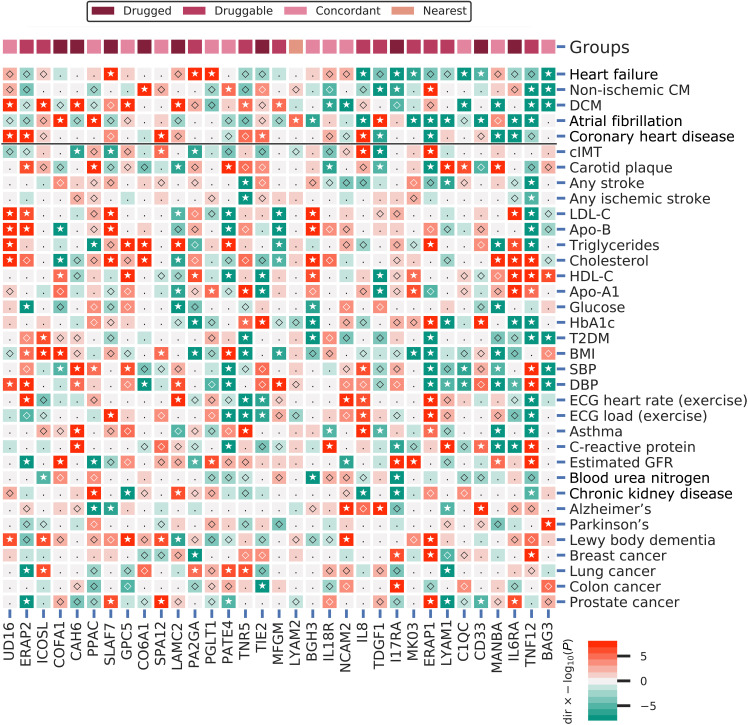
Fig. 6. A phenome-wide scan of CMR prioritized proteins associated with one or more cardiac outcomes.
Proteins were curated on having a multiplicity-corrected P < 1.29 × 10−5 with one or more of the following cardiac traits: HF, DCM, non-ischemic CM, AF, or CHD. P values passing the 0.05 threshold are indicated by an open diamond, with stars indicating results passing a threshold of 1.29 × 10−5. Cells were colored by effect direction multiplied by the −log10(P value); where P values were truncated at 8 for display purposes. The top column indicates whether the CMR-associated proteins were identified as drugged, druggable, directionally concordant, or nearest druggable protein. DCM, dilated cardiomyopathy; cIMT, carotid artery intima media thickness; T2DM, type 2 diabetes; BMI, body mass index; DBP/SBP, diastolic/systolic blood pressure; Estimated GFR, estimated glomerular filtration rate; BUN, blood urea nitrogen; LDL-C, low-density lipoprotein cholesterol; HDL-C, high-density lipoprotein cholesterol; Apo-B, apolipoprotein-B; Apo-A1, apolipoprotein-A1; HbA1c, glycated hemoglobin; ECG, electrocardiography; FVC, forced vital capacity; FEV1, forced expiratory volume during the first second; PEF, peak expiratory flow. Note that all 56 phenome-wide traits are presented in fig. S15. Nomenclature: Proteins are referred to by their UniProt entry name to differentiate them from the encoding genes.