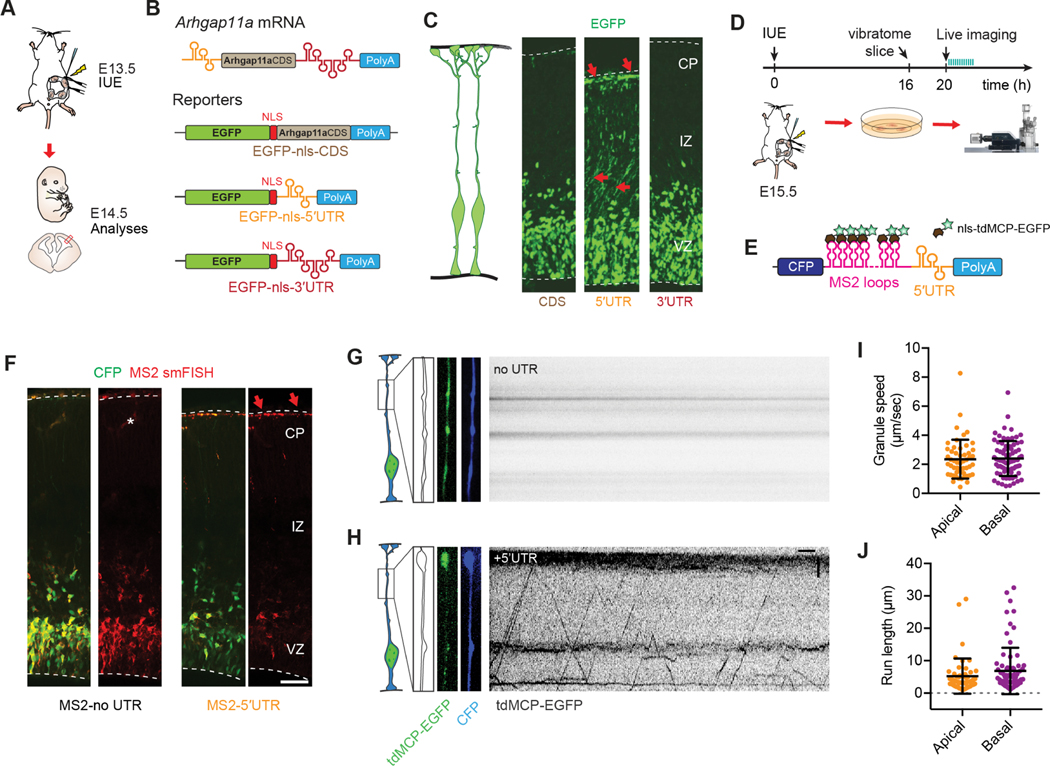
Figure 5. Arhgap11a mRNA is actively transported to radial glial basal endfeet via a 5′UTR element.
(A,B) Schematic overview (A) of the strategy used in (B, C) to determine the endfoot localization element in Arhgap11a mRNA using a 1-day electroporation of indicated reporter constructs (B). (C) EGFP-nls localizes to RGC basal endfeet only when the Arhgap11a 5′UTR is present, but not in CDS alone or containing 3′UTR.
(D, E) Schematic overview (D) of the strategy used in (F-J) to visualize transport of Arhgap11a mRNA reporters (E) in RGC basal processes.
(F) smFISH (red) targeting MS2 stem-loop RNA sequences shows Arhgap11a 5′UTR induces RNA localization from cell bodies to RGC basal process and endfeet (CFP, green).
(G,H) Kymographs showing absence (G) and presence (H) of MS2-tagged mRNA transport in RGC basal process over a 1-min period, in no UTR and 5′UTR, respectively.
(I,J) Quantification of similar average speeds of MS2-tagged mRNA transport in RGC basal processes (I) and average run lengths (J) in both apical and basally directed movements. n=126 EGFP+ punctae, 11 cells, 2 brains, 2 independent experiments.
IUE: in utero electroporation, CDS: coding sequence, UTR: untranslated region, nls: nuclear localization signal, tdMCP: tandem MS2-coat protein, CFP: cyan fluorescent protein, CP: cortical plate, IZ: intermediate zone, VZ: ventricular zone. Scale bars: F: 50μm, G,H: horizontal axis: 5sec, vertical axis: 5μm.