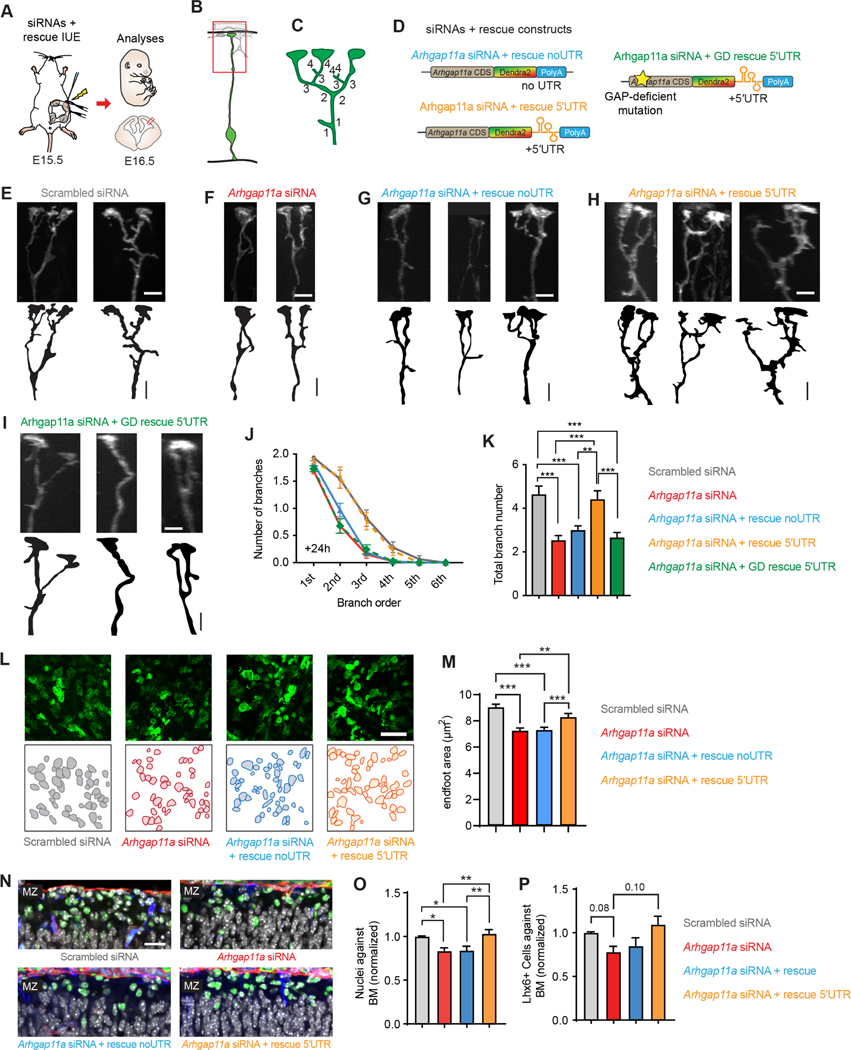
Figure 7. Locally synthesized ARHGAP11A controls basal process morphology through GAP activity.
(A) Schematic overview of the strategy used in (B-K) to assess rescue of RGC endfeet morphology.
(B,C) Method to define branch orders in RGC basal processes in MZ.
(D) Rescue constructs used in experiments.
(E-I) Representative images showing basal process complexity at the level of the MZ in RGCs treated as indicated.
(J,K) Quantification of basal process and endfoot complexity at the level of the MZ in RGCs. (Scrambled: n=78 cells, 6 brains, 4 independent experiments, Arhgap11a: n=78 cells, 5 brains, 4 independent experiments, Arhgap11a + rescue: n= 97 cells, 6 brains, 4 independent experiments, Arhgap11a + rescue 5′UTR: n= 56 cells, 4 brains, 3 independent experiments, Arhgap11a + GD rescue 5′UTR: n=78 cells, 4 brains, 2 independent experiments, J, 2-way ANOVA: p-value<0.0001, K, One way ANOVA: p<0.0001, Tukey’s Post-Hoc comparisons)
(L,M) Quantification of endfoot-basal lamina contact area in RGCs. (Scrambled: n=351 endfeet, 6 brains, 4 independent experiments, Arhgap11a: n=351 endfeet, 6 brains, 4 independent experiments, Arhgap11a + rescue: n= 454 endfeet, 7 brains, 4 independent experiments, Arhgap11a + rescue 5′UTR: n= 200 endfeet, 4 brains, two independent experiments, Kruskal-Wallis test).
(N) Images of interneurons (LHX6, green) along the basement membrane (BM, red) in different genotypes.
(O, P) Quantification of the positioning of DAPI cells (white) and interneurons along the basement membrane. (Nuclei against BM: Scrambled: n=13 brains, 9 independent experiments, Arhgap11a: n=12 brains, 7 independent experiments, Arhgap11a + rescue: n=14 brains, 8 independent experiments, Arhgap11a + rescue 5′UTR: n= 13 brains, 8 independent experiments, One-way ANNOVA followed by Tukey Post-Hoc analyses; Lhx6+ cells against the BM: Scrambled: n=6 brains, 4 independent experiments, Arhgap11a: n=6 brains, 3 independent experiments, Arhgap11a + rescue: n=6 brains, 3 independent experiments, Arhgap11a + rescue 5′UTR: n= 8 brains, 4 independent experiments, Brown-Forsythe and Welch ANOVA followed by Dunnett T3 Post-Hoc analyses).
IUE: in utero electroporation, siRNA: small interfering RNA, CDS: coding sequence, UTR: untranslated region, GD: Rho-gap-deficient. **: p-value<0.01. ***: p-value<0.001. E-G: 5 μm. Graphs, average values +/− SEM.