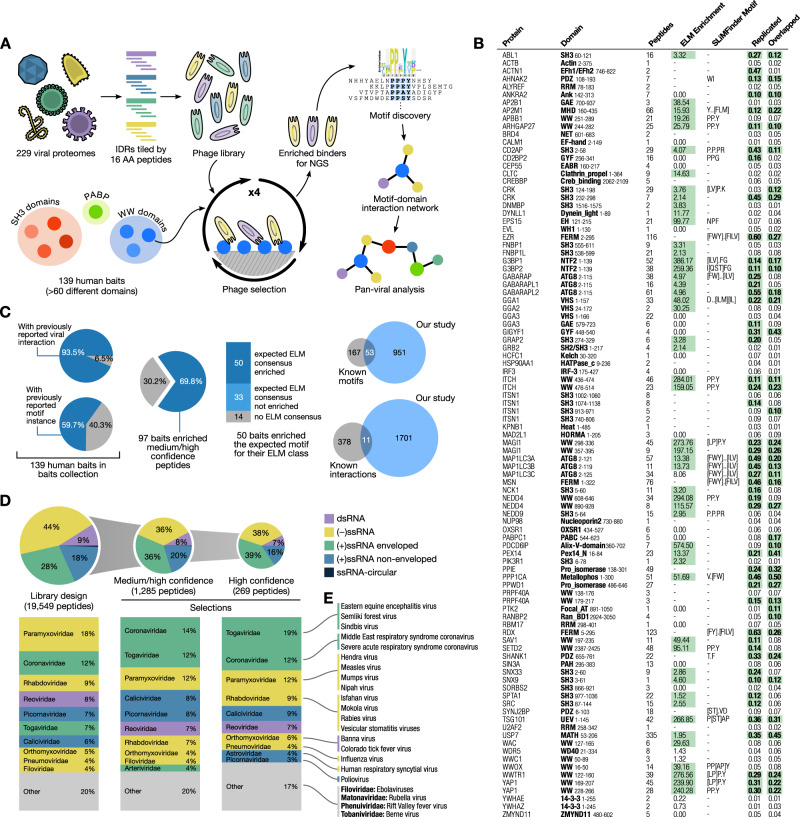
Fig. 1. Overview of the RiboVD library design and selection outcome.
A General workflow of the ProP-PD selection process and the data analysis. B Overview of the RiboVD selection results showing for each bait the number of enriched medium/high confidence peptides, the enrichment of peptides with sequences matching the consensus motif reported in ELM database, the generated motifs based on the enriched peptides (SLiMFinder Motif), and the quality of the selection results. “Replicated” represents the proportion of all peptides found in selections that were replicated in independent selections with the same bait while “Overlapped” is the proportion of all peptides that had overlapping peptides among all replicates for each bait. Green highlight indicates high quality of results. C Overview of the bait collection used in this study and the comparison of the selection results in context of previously known information. Left, percentage of baits used with previously known viral interactors (130 out of 139, Supplementary data 2) or previously reported motif instances (83 out of 139, Supplementary data 4). Center, percentage of bait domains that enriched peptides in selections and how their enriched motifs (if any) relate to the 83 motif instances previously reported in ELM. Right, overlap between the number of interactions in the RiboVD selections results and previously reported human-virus SLiM-based interactions (Supplementary data 4), or human-virus PPIs (Supplementary data 5). D RiboVD library composition and peptide distribution before and after selections. Represented as pie-plots are the percentages of RiboVD peptides belonging to the five different RNA viral genome types before (library design) and after selections. Below the pie charts are the top 10 viral families present in each pool. The representation of peptides for viruses with ssRNA-circular genomes was 103 peptides (0.5%) for the RiboVD library design, and 3 peptides (0.2%) for the medium/high confidence peptide set. E Examples of viruses from different virus families investigated in this study.