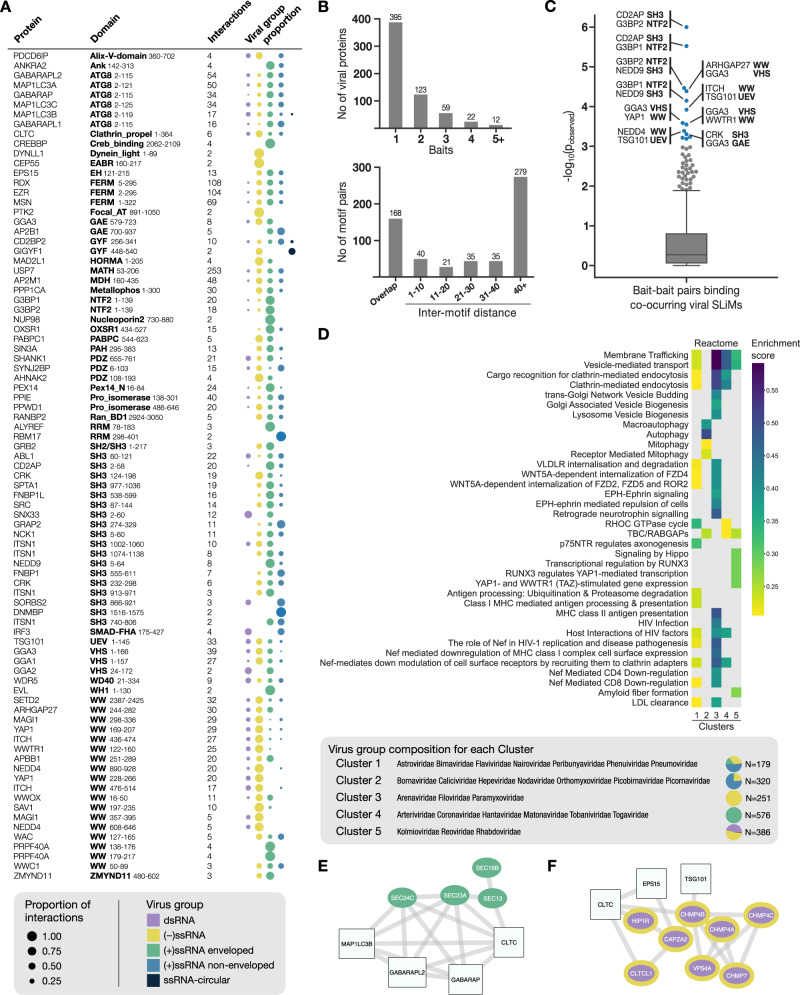
Fig. 2. Viral-host PPI and network analysis.
A Overview of the interactions identified per bait, together with the distribution of ligands from different types of viruses. B Viral targeting of bait proteins and motif proximity. Top: Number of screened baits recognized per viral protein showing that over 200 viral proteins bind to 2 or more human bait proteins. Bottom: Distribution of co-occurring viral motif-motif distances (in amino acids) showing that about a third of the viral peptides (208 out of 578) are 10 or less amino acids apart with most of them directly overlapping with each other. C Analysis of co-occurrence of host-binding peptide ligands in viral proteins (that is, peptides located in the same viral protein binding distinct bait domains) and their binding to protein domains. The p-values of the enrichment of co-occurring peptide ligands targeting human bait-bait pairs are shown (Supplementary data 6). Each dot represents a unique human bait-bait pair being targeted by co-occurring viral peptide ligands. Bait-bait pairs with p < 0.001 (based on randomization) are highlighted in blue and the top 10 human bait-bait pairs are labeled. D Clustering of host hijacking network signatures revealed five groups enriched in similar and distinct Reactome pathways. The relative frequency represents the enrichment score adjusted to account for the number of members in each viral family that are contributing to the enrichment. The N next to the pie charts indicates the number of identified interactions for all viruses in the cluster. E Sub-network of the COPII complex components (green) identified for cluster 4 based on network diffusion, together with their first neighbor bait proteins used in the RiboVD screen. F Sub-network of the ESCRTIII components (purple with yellow border) identified for cluster 5 based on network diffusion, together with their first neighbor bait proteins used in the RiboVD screen.