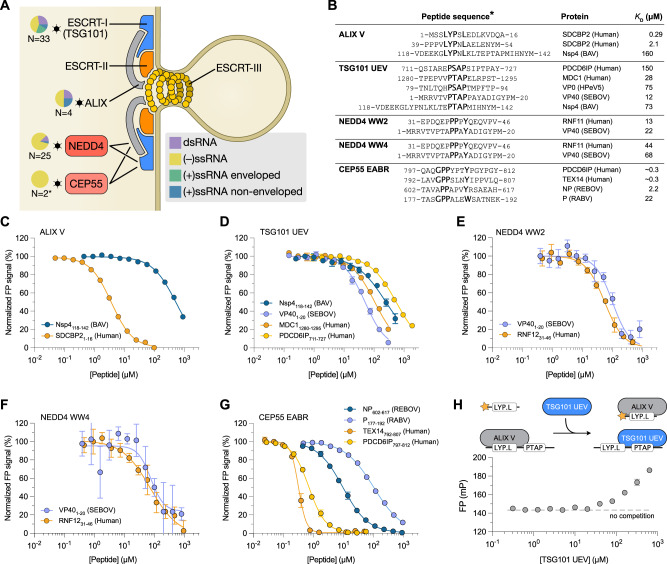
Fig. 3. The ESCRT machinery is hijacked by viral SLiMs that bind to NEDD4 WW, TSG101 UEV and ALIX V, and potentially also to CEP55 EABR.
A Schematic representation of the ESCRT pathway leading to reverse topology budding. Pie charts next to each target show the class of viral species hijacking it. N represents the number of identified interactions. B Overview of the peptides that bind to ESCRT pathway proteins for which affinities were measured. Residues constituting the recognition motif are shown in bold. C–G FP-monitored displacement experiments of viral and human peptides and ESCRT proteins. All FP-monitored experimental data in this paper are represented as normalized means ± SD of at least three replicate experiments. For detailed information on the peptides used in this study see Supplementary data 8. For all FP affinity measurements performed in this study see Figure S6. Source data for these and all subsequent FP-monitored measurements are provided in Source data files. H FP-monitored displacement experiment of Nsp4118-142 (BAV) shows that the interaction with ALIX V and TSG101 UEV is mutually exclusive.