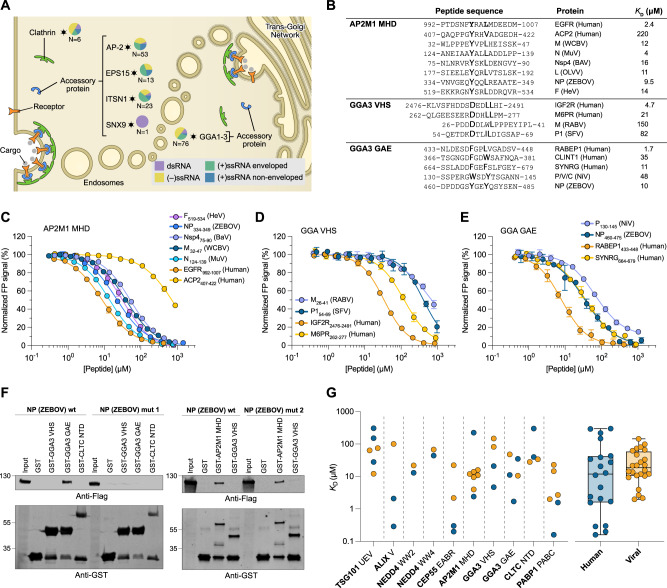
Fig. 4. Viral mimicry of distinct trafficking motifs binding to clathrin adapters.
A Schematic representation of clathrin adapter vesicle coat components for which viral ligands were found: AP2B1 and AP2M1 (collapsed as AP-2), CLTC, EPS15, ITSN1, SNX9, GGA1, GGA2, and GGA3 (collapsed as GGA1-3). Pie charts show the class of viral species hijacking the domain. N indicates the number of identified interactions. B Overview of affinity data for peptides interacting with clathrin adapter proteins. C–E FP-monitored affinity measurements of viral and human peptides and host proteins. Data are represented as normalized means ± SD of at least three replicate experiments. F Capture of full-length viral proteins by GST-tagged domains as visualized by Western blot. The interaction between NP (ZEBOV) and GGA3 GAE is lost upon motif mutation (Y469A). NP (ZEBOV) also interacts with AP2M1 MHD, but the introduced motif mutations (Y340A/L343A) did not abrogate binding suggesting additional AP2M1-binding SLiMs in NP (ZEBOV). Original blots for these and all subsequent Western blot experiments are provided in Source data files. G Overview of all affinity data for viral (orange) and host (blue) peptides generated in this study. In the right panel the combined data is presented as box plot showing mean, interquartile range, and minimum/maximum.