Figure 1. SHAPE-determined RNA secondary structures of wild-type PB2 and packaging mutant vRNAs.
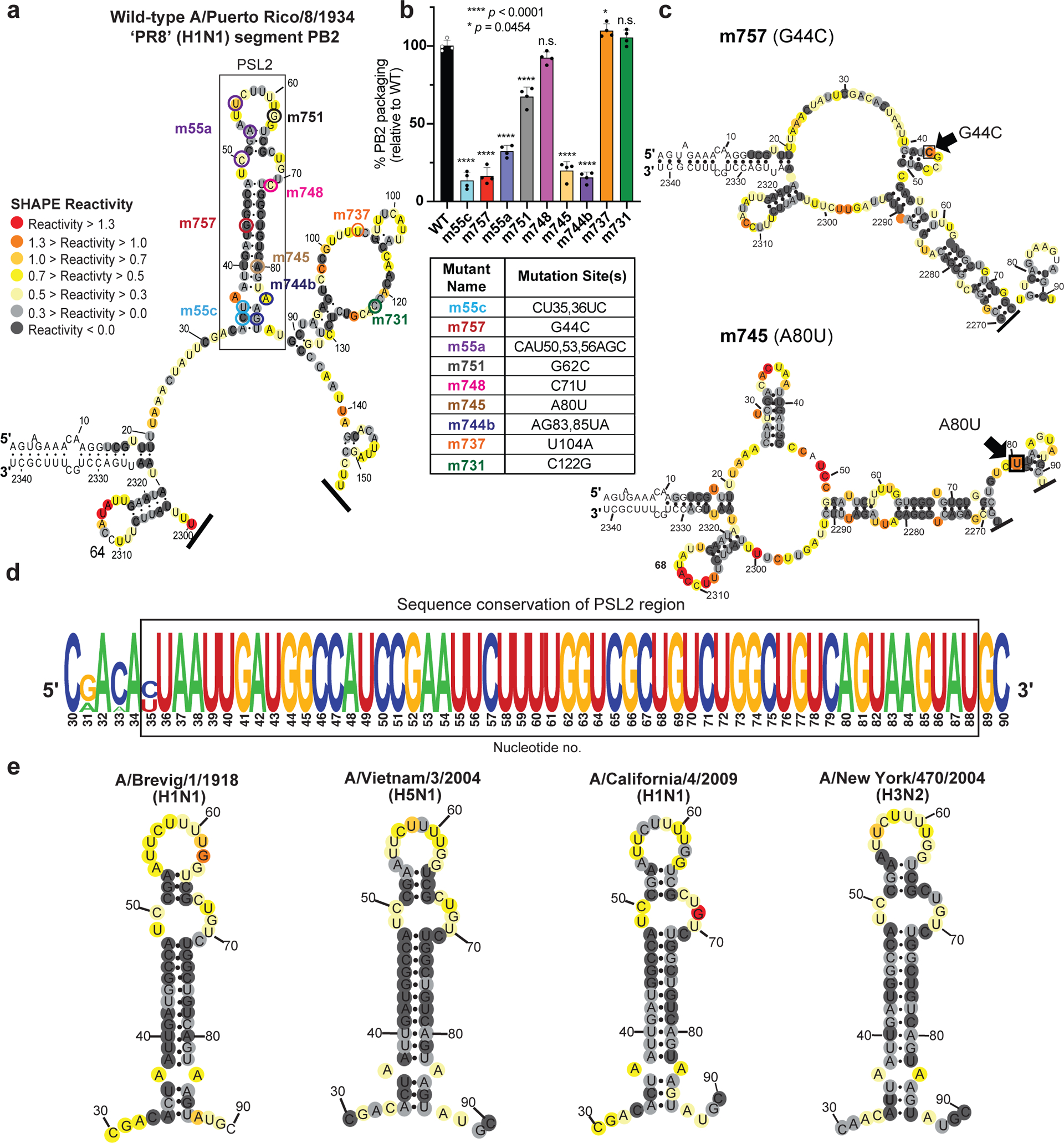
SHAPE-chemical mapping performed on full-length (−)-sense PB2 vRNAs. Colors denote SHAPE reactivity, which is proportional to the probability that a nucleotide is single-stranded. All structures are truncated to highlight the 5′ termini sequence structure. (a) SHAPE-predicted wild-type PB2 RNA secondary structure from strain A/Puerto Rico/8/1934 “PR8” (H1N1). Color-coded circles correspond to nucleotides sites where synonymous mutations were reported to affect PB2 packaging11,14. (b) Packaging efficiency of synonymous mutants in (a), determined by RT-qPCR. Results representative of two independent experiments with biological replicates, each performed in triplicate. Statistical analysis was performed using one-way ANOVA with Dunnett’s multiple comparisons test against the WT mean by GraphPad Prism 9 software (n=4). Error bars represents mean ± standard deviation (s.d.); **** p < 0.0001; * p = 0.0454, n.s. = not significant. Box below indicates mutant name and corresponding nucleotide change. Nucleotide numbering shown in the genomic (−)-sense orientation. (c) SHAPE-mapped structures of PB2 packaging-defective mutant vRNAs, m757 (G44C) and m745 (A80U) indicating loss of PSL2’s RNA secondary structure. Black arrowheads and boxed nucleotides denote site(s) of synonymous mutation. (d) Web-logo representation of the PSL2 region conservation across IAV strains and diverse influenza A viral subtypes (weblogo.berkeley.edu). The overall height represents sequence conservation at that nucleotide position, while the symbol height within each position indicates the relative frequency of each nucleotide at that site. Black box denotes PSL2 region. Sequences included in the alignment: pandemic A/Brevig Mission/1/1918 (H1N1), pandemic A/California/04/2009 (H1N1), seasonal human A/New York/470/2004 (H3N2), A/Puerto Rico/8/1934 (H1N1), highly pathogenic avian A/Vietnam/03/2004 (H5N1), avian A/mallard/Maryland/14OS1154/2014 (H6N1), pandemic A/Hong Kong/8/1968 (H3N2), and seasonal human A/New York/312/2001 (H1N1) (see Supplementary Fig. 1d). RNA nucleotides are numbered in (−) -sense orientation. (e) SHAPE-mapped structures of full-length wild-type PB2 vRNA from pandemic and highly pathogenic strains, including different subtypes to modern human strains: 1918 pandemic (A/Brevig Mission/1/1918 (H1N1)), highly-pathogenic avian (A/Vietnam/1203/2004 (H5N1)), 2009 pandemic ‘swine’ (A/California/04/2009 (H1N1)), and Fujian-like human seasonal virus, A/New York/470/2004 (H3N2)
