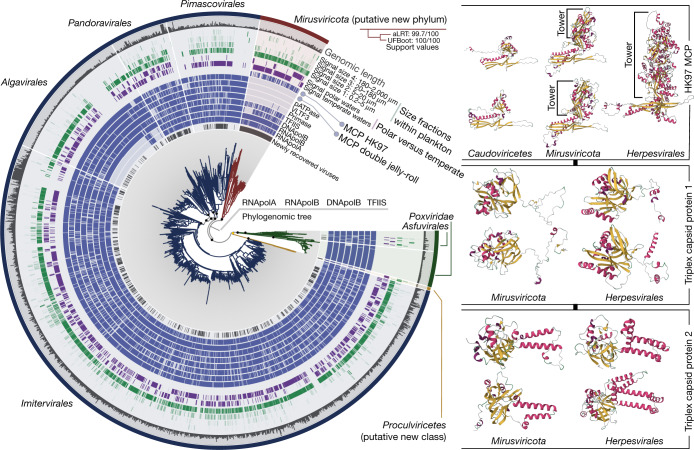
Fig. 1. Evolutionary relationships between Nucleocytoviricota, Herpesvirales and mirusviruses.
Left: a maximum-likelihood phylogenetic tree built from the GOEV database (1,722 genomes) on the basis of a concatenation of manually curated RNApolA, RNApolB, DNApolB and TFIIS genes (3,715 amino acid positions) using the posterior mean site frequency mixture model (LG + C30 + F + R10) and rooted between mirusviruses and the rest. Highlighted phylogenetic supports (dots in the tree) were considered high (approximation likelihood ratio (aLRT) ≥ 80 and ultrafast bootstrap approximation (UFBoot) ≥ 95, in black) or medium (aLRT ≥ 80 or UFBoot ≥ 95, in yellow; see Methods). The tree was decorated with rings of complementary information and visualized with anvi’o. Right: predicted 3D structures for the HK97 MCP of Caudoviricetes, mirusvirus and herpesvirus representatives obtained using AlphaFold2. Proteins are coloured on the basis of secondary structure properties. The panel also shows predicted 3D structures for the triplex capsid proteins of mirusvirus and herpesvirus representatives using the same methodology.