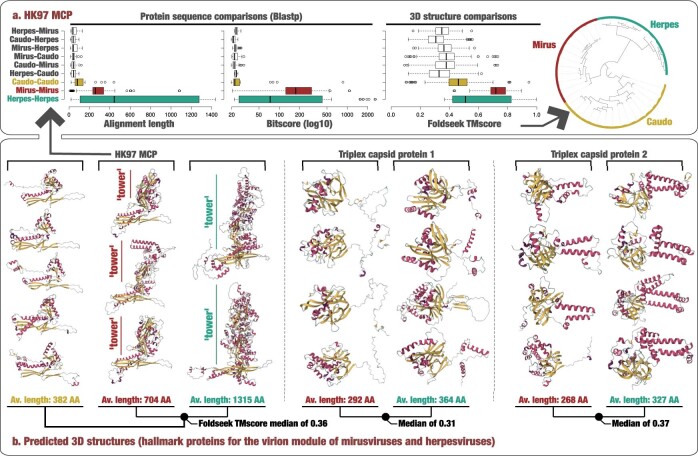
Extended Data Fig. 4. Protein sequence and predicated 3D structures comparisons.
Panel A displays protein sequence and 3D structure comparisons (Blastp and Foldseek) for the HK97 MCP of representatives covering various families from the three main Duplodnaviria clades. Center lines in boxplots show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles; outliers are represented by dots (from top to bottom, n = 22, 50, 38, 25, 35, 16, 40, 23 and 117 independent comparisons). The alignment values range from a minimum of 9 amino acids to a maximum of 1,437 amino acids. The bitscore values range from a minimum of 19.6 to a maximum of 2577. The Foldseek TMscore values range from a minimum of 0.09 to a maximum of 0.997. The dendrogram was generated using Euclidian distance and ward within anvi’o and is based on the Foldseek TMscore values. Panel B describes a selection of predicated 3D structures for the HK97 MCP and triplex proteins of representatives from the three main Duplodnaviria clades (Caudoviricetes viruses lack the triplex capsid proteins). Proteins are colored based on secondary structure properties.