Abstract
Analysis of premalignant tissue has identified the typical order of somatic events leading to invasive tumors in several cancer types. For other cancers, premalignant tissue is unobtainable, leaving genetic progression unknown. Here, we demonstrate how to infer progression from exome sequencing of primary tumors. Our computational method, PhylogicNDT, recapitulated the previous experimentally determined genetic progression of human papillomavirus-negative (HPV–) head and neck squamous cell carcinoma (HNSCC). We then evaluated HPV+ HNSCC, which lacks premalignant tissue, and uncovered its previously unknown progression, identifying early drivers. We converted relative timing estimates of driver mutations and HPV integration to years before diagnosis based on a clock-like mutational signature. We associated the timing of transitions to aneuploidy with increased intratumor genetic heterogeneity and shorter overall survival. Our approach can establish previously unknown early genetic progression of cancers with unobtainable premalignant tissue, supporting development of experimental models and methods for early detection, interception and prognostication.
Subject terms: Cancer genomics, Head and neck cancer, Computational biology and bioinformatics, Cancer
Getz and colleagues demonstrate a computational approach to estimate the timing of genomic drivers in early tumor progression from cancer types that do not have premalignant disease, using HPV-negative and HPV-positive head and neck squamous cell carcinoma as examples.
Main
Deciphering the temporal order of genetic lesions throughout the steps of cancer progression has long been a goal of cancer research1,2. That order can provide clues to etiology and cell-intrinsic mechanisms in tumorigenesis, informing studies of how normal tissue becomes a tumor, and can also provide ways to detect and treat early disease stages and identify early clonal events as promising targets for therapy of later invasive tumors3.
For cancers with well-defined pathologic progression from normal tissue, for example, adenoma through carcinoma in situ to invasive carcinoma, analysis of lesions along that trajectory has provided corresponding genetic progression models4–13. However, there are many cancer types with poorly defined, undetectable or difficult-to-biopsy premalignant lesions14–22 whose genetic progression thus remains speculative.
Here, we demonstrate how to infer the typical order of genetic events in a cancer type from exome sequencing of primary tumor samples taken long after cancer initiation. The genome of an invasive tumor includes information about its initial genetic progression, which is recorded in patterns of somatic mutations and copy number changes as cells evolve into an invasive clone23,24. We recently developed PhylogicNDT, an integrated suite of tools, to reconstruct the clonal architecture and relative timings of genetic events using a coherent probabilistic framework25,26 and have successfully applied it in several contexts25,27–29.
We asked whether more readily available whole-exome sequencing (WES) of primary tumors could similarly provide information on genetic progression.
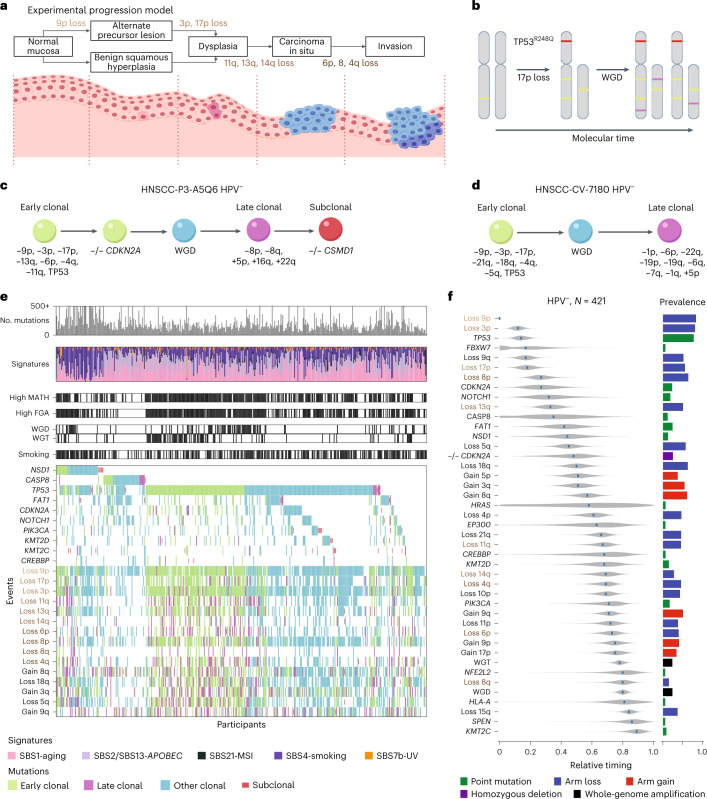
We examined head and neck squamous cell carcinoma (HNSCC), which provides a unique opportunity to validate and extend such progression models. A quarter of a century ago, Califano et al.8 determined the genetic progression of classic HNSCC, typically associated with tobacco and alcohol use, from loss-of-heterozygosity (LOH) analysis along the pathologic progression from normal tissue to initial lesion, dysplasia and carcinoma in situ to invasive carcinoma (Fig. 1a). That model, as confirmed and extended to other genetic events in subsequent studies30–34, continues to provide the framework for HNSCC progression35,36. A valid computational reconstruction of genetic progression from WES of primary HNSCC should agree with those findings and provide timing for additional genetic events.
Fig. 1. Genetic events and their timing in HPV– HNSCC.
a, Diagram of HPV– genetic progression determined by Califano et al.8 from tissue samples taken near the tissue surface at different stages of disease progression with associated genetic events (left to right). b, Conceptual basis of estimating timing within tumors. The loss of chromosome arm 17p most likely occurred before the WGD event. The mutation TP53R248Q most likely occurred before WGD, based on estimated multiplicity. Additional mutations (purple) are used to estimate the mutational relative timing of the WGD event. c,d, Examples of timing of genetic events in individual tumor samples; –, chromosome arm loss; +, chromosome arm gain; –/–, homozygous deletion in the indicated gene. e, CoMut plots for 421 HPV– HNSCCs. For each tumor, from top to bottom, the number of mutations, their mutational signatures, MATH, FGA, the presence of whole-genome amplification (WGD and WGT), smoking within 15 years of diagnosis and selected genetic events are shown. The shading corresponds to timing of individual events. MSI, microsatellite instability. f, Relative timing of genetic events based on 421 HPV– HNSCCs. The analysis compared 43 events among tumors: whole-genome amplification (WGD and WGT), arm losses examined by Califano et al.8 (names are colored with respect to timing groups in a) and other events with notable prevalence among HPV– tumors (Methods). Events are ordered top to bottom by the point estimates of their mRT scores. Violin plots illustrate the posterior distributions of relative timing. The event prevalence and type (color coded) are displayed to the right of the corresponding violin plot.
In contrast to classic HNSCC, the genetic progression of the increasingly prevalent and clinically important HNSCC associated with human papillomavirus (HPV;37,38 abbreviated HPV+ HNSCC) has remained unknown39. Premalignant lesions have not been identified reliably for HPV+ HNSCC39,40. The early genetic events in classic HPV– HNSCC that disrupt the CDKN2A and TP53 loci are infrequent in HPV+ HNSCC, as the E7 and E6 viral protein products provide corresponding tumorigenic functions41,42. Whether HPV+ HNSCCs converge thereafter to the same genetic progression as HPV– HNSCC is unclear. Computational reconstruction provides a unique opportunity to determine the genetic progression of HPV+ HNSCC and explore whether the well-known differences between HPV+ and HPV– HNSCC in response to therapy35 are associated with their different genetic paths to invasive cancer.
We thus applied PhylogicNDT to WES data from primary HNSCC to (1) demonstrate the reconstruction of genetic progression in HPV– HNSCC and (2) identify the genetic progression of HPV+ HNSCC. We started with HNSCC from The Cancer Genome Atlas (TCGA) project43 and collected samples from 43 additional oropharyngeal HNSCCs to increase the representation of HPV+ cases (101 total HPV+ HNSCCs).
Here, we report the validation of our computational approach in HPV– HNSCC, extending the number and details of timed events, and describe the previously unknown order of genetic progression of HPV+ HNSCC. We document the timing of different types of aneuploidy and their strong association with intratumor heterogeneity and survival. Additionally, we converted the relative timing estimates of main drivers and HPV integration to years before diagnosis based on a clock-like mutational signature.
This report provides a framework for studying genetic progression of cancer types for which premalignant tissue is unobtainable. This framework enables uncovering mechanisms of cancer initiation and early development in types of cancer for which the rarity, inaccessibility, or lack of premalignant tissue previously made their genetic progressions undecipherable.
Results
The data in Fig. 1b show how the order of genetic events during tumor development is inferred from a tumor sample obtained long after initiation, once the fraction of cancer cells that harbor each mutation (cancer cell fractions (CCFs)) and their multiplicities (the number of DNA molecules carrying a mutation per cancer cell) are determined. In this example, cancer cells in the final tumor sample (Fig. 1b, right) showed four copies of chromosome arm 17q but only two copies of arm 17p, consistent with LOH at 17p followed by duplication. The final pattern of somatic mutations (colored bands) was consistent with some mutations (green) before the duplication and others (purple) thereafter. The driver mutation in TP53 (red) leading to a R248Q protein alteration was seen in both final copies of arm 17p, consistent with the mutation preceding the duplication.
Relative timing of all genetic events within each tumor is estimated across the genome by applying these principles in a probabilistic computational framework25,26. Examples of inferred orders of events in two HPV– tumors are shown in Fig. 1c,d. The inferred timing of each event is represented as a posterior distribution over a molecular mutational time scale π, with π = 0 and π = 1 reflecting the times of the first and last clonal events, respectively26.
Next, PhylogicNDT combines the within-tumor timing (π value distributions) of selected driver events across a cohort of tumors to obtain a posterior distribution for the typical relative timings of the events. The median relative timing (mRT) for each genetic event provides a point estimate for ordering events into a genetic progression model.
We applied PhylogicNDT to WES data of 531 HNSCC tumor–normal pairs, with 421 HPV– and 101 HPV+ tumors. We focused on 64 established or candidate HNSCC drivers, including 24 frequently mutated genes and 40 frequently gained or lost genomic regions in either or both HPV– and HPV+ tumors (Supplementary Tables 1 and 2).
Corroborating timing of driver events in HPV– HNSCC
The genomic landscape of 421 HPV– tumors is illustrated in Fig. 1e. For each participant’s tumor, we report the number of mutations, relative activity of different mutational signatures, measures of genetic heterogeneity, smoking history and the occurrence of the most frequent 10 driver genes and 15 copy number alterations, with tumor-specific timing estimates.
We computationally inferred the order of events among these HPV– HNSCCs and compared the order to the empirically derived progression model of Califano et al.8 (Fig. 1a). In that model, preneoplastic tissue arises by loss of chromosome arm 9p (−9p; where CDKN2A resides), progressing to dysplasia with −17p (which includes TP53) and −3p. The transition to carcinoma in situ is associated with −13q, −11q and −14q, and, finally, progression to invasive cancer is associated with −6p, −8 and −4q. Even the partial orderings of within-tumor timing estimates were generally consistent with the Califano model (Fig. 1b–e). We combined the participant-specific orders of events across the cohort (using the PhylogicNDT LeagueModel tool; Methods) to obtain the typical order of the 43 most prevalent driver events (Fig. 1f).
Computational timing analysis, based solely on primary tumor specimens, recapitulated and substantially expanded the empirical progression model of Califano et al. (Fig. 1a). Notably, losses described by Califano et al. appeared in our model in the same order as in their model. We predicted that timings of losses of the p and q arms of chromosome 8 are different (P = 6.2 × 10−5). Indeed, the early loss of arm 8p, not evaluated by Califano et at., agreed with its frequent loss in early oral cavity dysplasia31.
The first and most prevalent event was –9p (prevalence of 84.6%; mRT = 0.05), the location of the CDKN2A tumor suppressor that is also inactivated by frequent (23.5%) and early (mRT = 0.27) point mutations or homozygous deletions (25.4% prevalence; mRT = 0.48). Nearly all HPV– HNSCC (388/421; 92.2%) had at least one such disruption of CDKN2A.
Disruption of TP53 was also frequent and early via mutation (78.4%; mRT = 0.14) or −17p (54.4%; mRT = 0.18), with at least one such event in 369 HPV– tumors (87.6%). Our timing of TP53 and CDKN2A single-nucleotide variations (SNVs) agreed with the long-appreciated presence of TP53 (ref. 30) and CDKN2A (ref. 32) mutations in dysplastic tissue of the head and neck. The third of the earliest genetic events identified by Califano et al., −3p, occurred in 339 tumors (80.5%); its mRT of 0.12 was similar to that of TP53 disruptions.
We further identified prevalent and early −9q (51.5%; mRT = 0.17) and −8p (62.9%; mRT = 0.27) events. By contrast, the frequent +3q event (55.6%), containing the PIK3CA locus, was later (mRT = 0.51), suggesting a role in progression rather than initiation. Other arm-level gains and losses covered a wide range of relative timings.
Based on the Califano et al. model, we mapped relative timings to pathologic progressions. Precursor lesions become dysplastic near an mRT of ~0.25, between timings of −17p (0.18) and −13q (0.33). The transition to carcinoma in situ is near 0.7 (mRT of −14q and −4q) and that to invasive carcinoma at or after ~0.75 (−6p mRT = 0.73; −8q mRT = 0.8). Our timing of +3q (mRT = 0.51) and +8q (mRT = 0.57) before this estimate of the transition to carcinoma in situ agrees with findings (published after the Califano model) of these gains in dysplastic tissue33,34.
We further established relative timing of SNVs in several lower-prevalence driver genes (in order of increasing mRT): FBXW7, NOTCH1, CASP8, FAT1, NSD1, HRAS, EP300, CREBBP, KMT2D, PIK3CA, NFE2L2, HLA-A, SPEN and KMT2C. mRT values of events in the first nine genes were less than 0.69, supporting early roles in tumorigenesis before carcinoma in situ. Although individually of low prevalence (each <25%), a mutation in at least one of these first nine genes occurred in over 60% of HPV– HNSCCs (260/421). Mutations in PIK3CA, NFE2L2, HLA-A, SPEN and KMT2C were at later mRT values, presumably late in pathologic progression.
Finally, whole-genome events, leading to whole-genome copy number profiles that are predominantly triploid (WGT) or whole-genome doubling of both alleles (WGD) resulting in tetraploid samples, occurred in nearly half (47.7%) of HPV– HNSCCs. These whole-genome events were late in progression (mRT values of 0.78 (WGT) and 0.80 (WGD); Fig. 1f).
Establishing the genetic progression of HPV+ HNSCC
Encouraged by our success with classic HPV– HNSCC, we turned to HPV+ HNSCC, where pathologic progression has not been identified35,44. These tumors typically arise in crypts of tonsils and related lymphoid tissues at breaks of the basement membrane35,45,46 that allow viral access to infect basal epithelial cells and ready entry of transformed cells into the lymphatics (Fig. 2a). HPV+ HNSCC often spreads to local lymph nodes before a primary tumor is identified; premalignant tissue has seldom been found35,47.
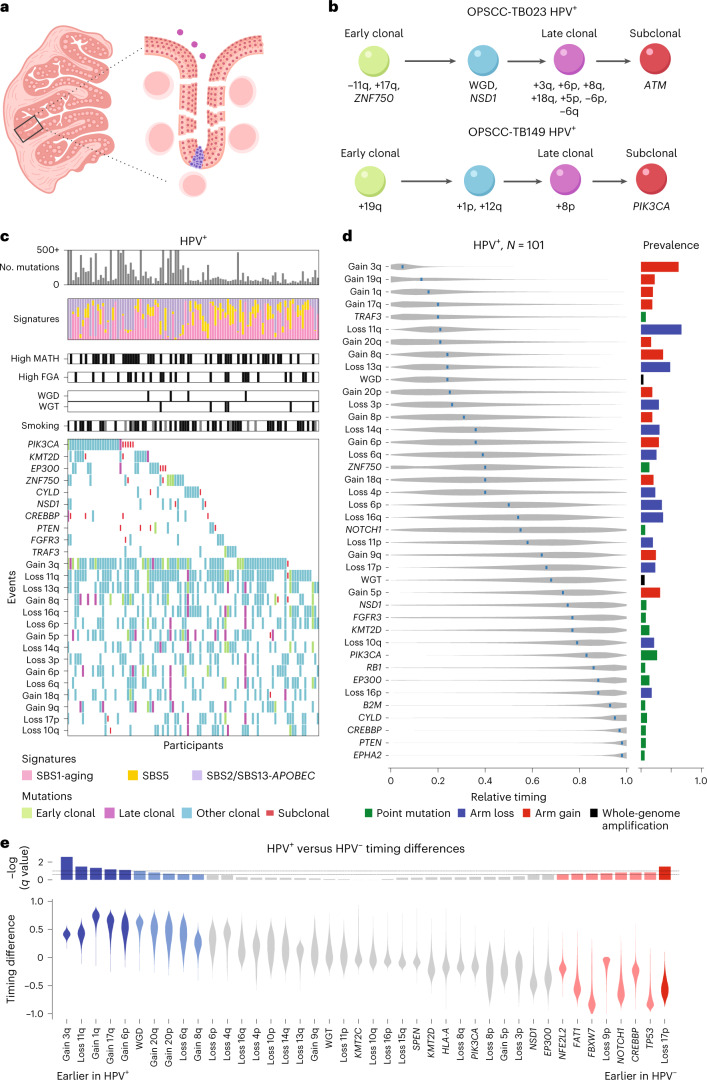
Fig. 2. Genetic events and their timing in HPV+ HNSCC.
a, Illustration of lack of sampling access and ease of tumor invasion with HPV+ tumors. Left, sketch of tonsil cross-section showing crypts; right, magnification of the base of a crypt illustrating entry of HPV (purple circles) and subsequent tumor development (blue) at a break in the discontinuous basement membrane. b, Examples of genetic event timing within individual tumor samples, represented as in Fig. 1c; OPSCC, oropharyngeal squamous cell carcinoma. c, CoMut plots for HPV+ HNSCC, represented as in Fig. 1e. The shading corresponds to timing from individual times with PhylogicNDT. The top 10 SNVs and 15 copy number variations by prevalence were selected for display. d, Relative timing of genetic events based on 101 HPV+ HNSCCs, represented as in Fig. 1f. The analysis compared 40 events among HPV+ tumors: whole-genome amplification (WGD and WGT), arm losses with >15% prevalence and SNVs with >5% prevalence. e, Relative timing between HPV classes of 42 shared events. Violin plots show the distributions of difference in HPV-class-specific timing. P values were derived from the MCMC posterior, as described in the Methods. Associated log10 (q) values are displayed at the top; N = 101 individuals with HPV+ HNSCC (b–d).
We assembled WES data from 101 HPV+ HNSCCs, including 65 from TCGA43 and 36 collected for this study. Examples of within-tumor timing are shown in Fig. 2b and Extended Data Fig. 1; the genetic landscape of HPV+ HNSCCs is illustrated in Fig. 2c. We found four mutational signatures in this cohort, with 28% of mutations attributed to aging signatures (single-base substitution 1 (SBS1) and SBS5) and 62% to APOBEC signatures (SBS2 and SBS13). Median tumor purity was 0.48 (range of 0.12 to 0.98), and median ploidy was 2.14 (range of 1.60 to 5.56; Methods).
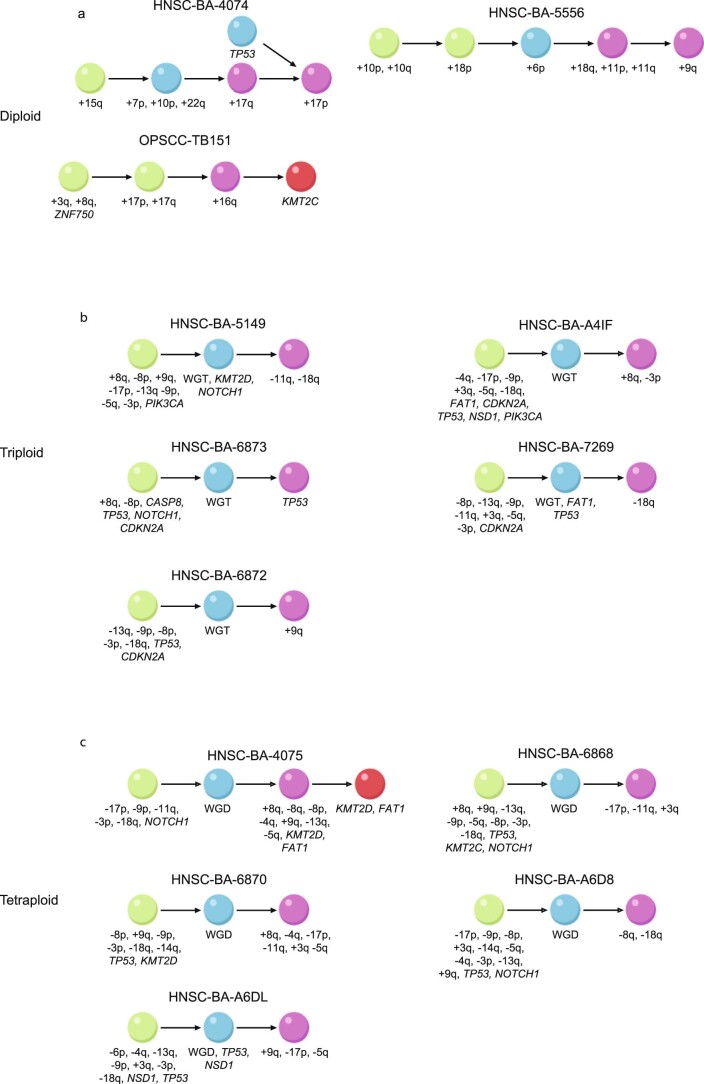
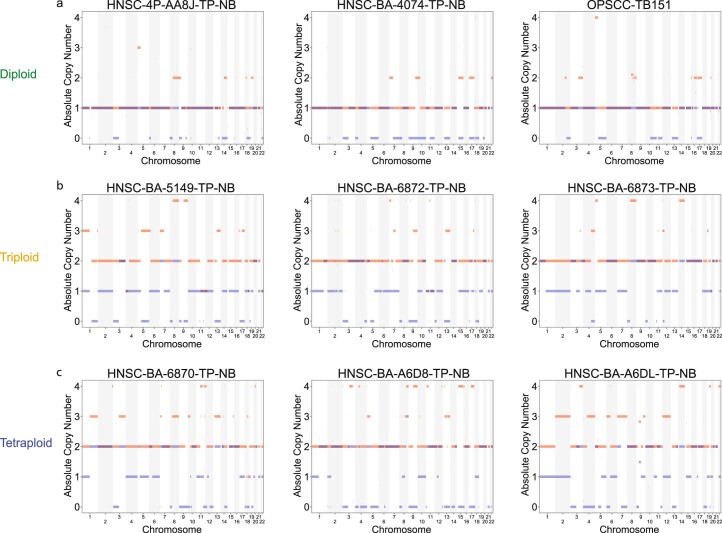
Extended Data Fig. 1. Single-patient timing.
Examples of genetic event timing within individual tumor samples classified as diploid (a), triploid (b), and tetraploid (c). WGT, whole-genome triploidy; WGD, whole-genome duplication; –, chromosome arm loss; +, arm gain; other symbols, mutation in the indicated gene. The colors of the balls represent the timing. Green, early clonal; purple, late clonal; blue, other clonal; red, subclonal.
As for HPV– HNSCC, we estimated the order of events (π) for each HPV+ tumor via PhylogicNDT SinglePatientTiming (Fig. 2c). We then combined timing information for 40 genetic driver events across tumors to infer the typical order in HPV+ cancers (Fig. 2d).
Gain of chromosome arm 3q and loss of 11q were both highly prevalent and early (+3q: 70.3% prevalence and mRT = 0.05; −11q: 69.3% prevalence and mRT = 0.20). Arm-level events −13q and +8q were also both highly prevalent (>40%) and early (mRT values of 0.21 and 0.24, respectively), suggesting frequent roles in early HPV+ progression. All other arm-level events had a prevalence of <40%.
Of 13 low-prevalence potential driver genes, mutations only in TRAF3, ZNF750 and NOTCH1 had mRT values of 0.55 or earlier, while others had mRT values of 0.75 or greater. Except for PIK3CA (mRT = 0.83), none of the 13 potential driver genes were mutated in more than 15% of HPV+ tumors, although nearly three-quarters of tumors (74/101, 73%) had a mutation in at least 1 potential driver gene. Only 10 of 101 HPV+ HNSCCs had genome-wide events leading to WGT or WGD.
Comparing HPV+ and HPV– genetic progressions
In HPV+ HNSCC, the HPV E7 and E6 viral gene products inactivate the functions of CDKN2A and TP53, whereas in HPV– HNSCC, those functions are lost via somatic events41,42. We sought to determine other similarities and differences in the progression of these two major HNSCC subtypes.
First, we compared the frequencies of genomic events in HPV+ and HPV– HNSCC. Even beyond expected differences associated with CDKN2A and TP53, 20 genetic events were significantly more prevalent in HPV– HNSCC, including SNVs in FAT1, NOTCH1, CASP8, HRAS, the −3p highly prevalent in HPV– HNSCC and 15 other arm-level somatic copy number alterations (Supplementary Tables 1 and 2; Fisher’s exact test with Benjamini–Hochberg false discovery rate q value of <0.1). Notably, whole-genome events leading to WGT or WGD were nearly five times more prevalent in HPV– (201/421, 48%) than in HPV+ tumors (10/101, 10%; P = 10−13, Fisher’s exact test). However, 14 events were found at higher prevalence in HPV+ than in HPV– HNSCC (SNVs in PIK3CA, ZNF750, EP300, CYLD, TRAF3, FGFR3, PTEN, B2M and RB1 and arm-level events +3q, −11q, −16q, −18q and +19q).
Comparing mutational signatures showed that the prevalence of all signatures differed between HPV+ and HPV– tumors, with APOBEC (SBS2/SBS13) and aging signatures (SBS1/SBS5) enriched in HPV+ tumors and the smoking signature (SBS4) enriched in HPV– tumors.
Next, to compare the timing of events, we examined 42 events present in at least three cases in each of the HPV+ and HPV– cohorts (Fig. 2e). We combined all anatomic sites, as the few (33) HPV– oropharyngeal tumors did not allow reliable anatomic site-specific timing comparisons. We calculated distributions of differences in relative timing and assessed statistical significance by permutation (Methods). We detected 6 differentially timed events at q < 0.1 (5 earlier in HPV+ and 1 in HPV–) and 12 more at q < 0.2.
Early high-prevalence genetic events in HPV– progression, −9p, −17p and mutation of TP53 (CDKN2A or TP53 inactivation), were earlier than in HPV+ tumors, as expected from the different mechanisms for inactivating the function of these tumor suppressor genes. Lower-prevalence mutations in CREBBP, NOTCH1, FBXW7, FAT1 and NFE2L2 were also earlier in HPV– tumors.
The early and high-prevalence genetic events in HPV+ progression, +3q and −11q, were significantly earlier in HPV+ than in HPV– progression, supporting their roles as early HPV+ driver events. WGD in HPV+ HNSCC was also found to occur significantly earlier than in HPV– tumors, despite the much lower prevalence of WGD in HPV+ tumors. Other arm-level events earlier in HPV+ progression were gains +1q, +17q, +6p, +20p, +20q and +8q and loss of 6q.
Progressions in HNSCC subclasses and at anatomic subsites
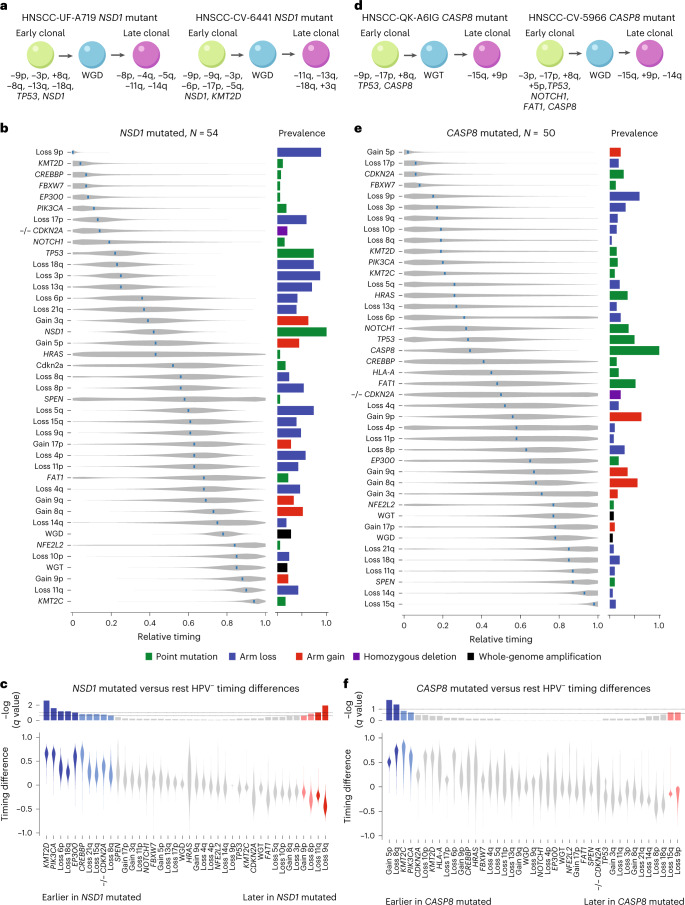
Among HPV– HNSCC, prevalence of some genetic events differed markedly among the major anatomic subdivisions oral cavity, oropharynx and larynx. NSD1 mutations were preferentially laryngeal, while almost all CASP8 mutations were in oral cavity tumors (Supplementary Table 3). That led to the question of whether tumors with these mutations, reflecting distinct anatomic sites, have different genetic progression trajectories. We used PhylogicNDT to infer genetic progression of HPV– HNSCC subsets with mutations in NSD1 (n = 54) or CASP8 (n = 50), comparing their timings against other HPV– tumors (Fig. 3). We also estimated timing in NOTCH1-mutated tumors (n = 54), which showed no significant subsite preference (Extended Data Fig. 2a–c).
Fig. 3. Timing within subsets of HPV– HNSCC.
a, Examples of single-tumor timing diagrams for tumors with mutations in NSD1. b, Relative timing diagram for 54 HPV– tumors with mutations in NSD1, represented as in Figs. 1f and 2d. c, Comparison of event timing between the NSD1-mutated and the remaining HPV– HNSCCs, represented as in Fig. 2e. d–f, Corresponding example of single-tumor timing diagram (d), relative timing diagram (e) and comparison of event timing diagram (f) for 50 tumors with mutations in CASP8.
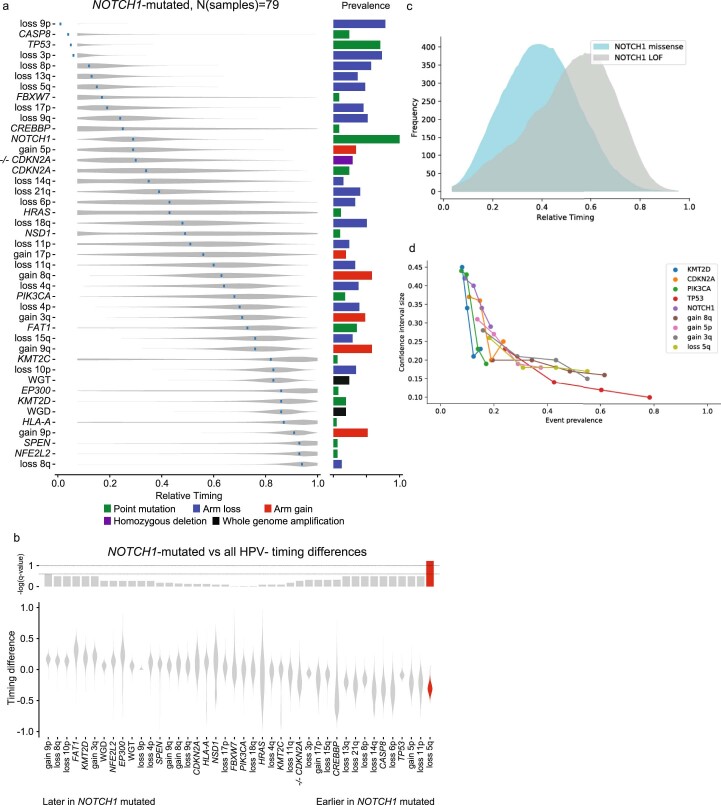
Extended Data Fig. 2. Timing within the NOTCH1-mutated subset of HPV- HNSCC.
(a) Relative Timing of genetic events, based on 79 NOTCH1-mutated HPV-negative HNSCCs. Analysis compared 43 events among tumors: whole-genome amplification (WGD, doubling; WGT, triploidy) and other events with notable prevalence among HPV-negative tumors (arm losses with > 30% prevalence, SNV with > 5% prevalence). (b) Relative timing between NOTCH1-mutated and all HPV-negative tumors of 42 events (all events from panel a excluding NOTCH1) (N = 79 NOTCH1-mutated patients). Violin plots show the distributions of difference in timing between the groups. Associated log10 q-values are displayed at the top. (c) MRT timing profiles for NOTCH1 activating and deactivating mutations (d) Power analysis through down sampling of higher prevalence events, demonstrating dependence of the confidence interval size on event prevalence (N = 421 HPV- patients).
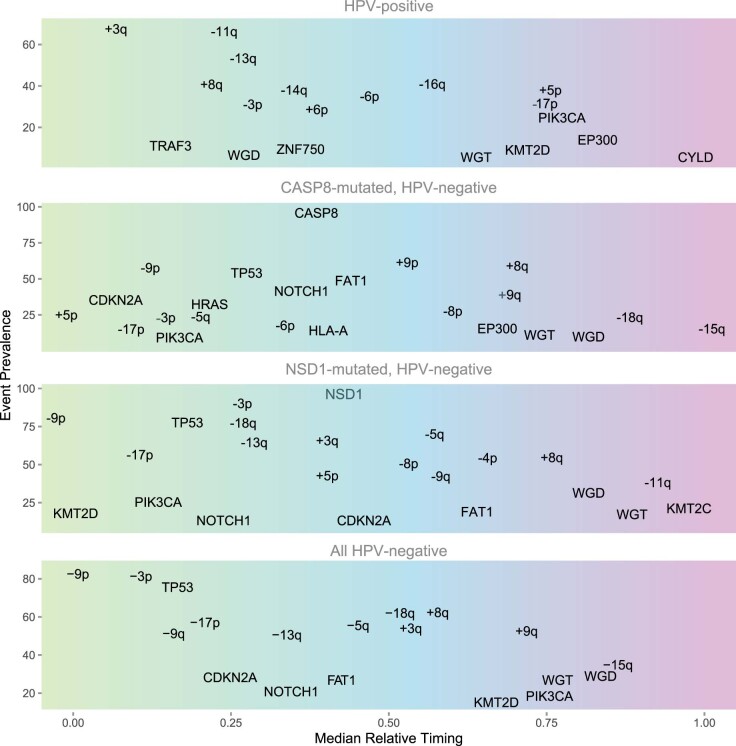
The preferentially laryngeal NSD1-mutated tumors (Fig. 3a–c) had several events whose timing differed from other HPV– HNSCCs, despite typically similar event prevalence between those two groups (Supplementary Table 4). Mutations in KMT2D, CREBBP, EP300 and PIK3CA timed at an mRT of ≤0.11 in NSD1-mutated cases versus at an mRT of ≥0.63 in other HPV– HNSCCs. Homozygous deletions of CDKN2A were also significantly earlier in NSD1-mutated tumors. NSD1 mutations themselves had an mRT of 0.42. Significantly later events were arm-level losses or gains. Some timing differences could be associated with increased smoking signature (SBS4) activity in NSD1-mutated tumors (Fig. 1b).
By contrast, CASP8-mutated tumors, almost solely in the oral cavity, showed major differences in event prevalence from other HPV– HNSCCs (Supplementary Table 5), while mRT values of most events were similar (Fig. 3e–f). Compared to other HPV– HNSCCs, they had a substantially lower prevalence of whole-genome events (14% versus 52.3%; P = 10−7, Fisher’s exact test). Nearly two-thirds of driver events (46 of 65) had differential prevalence (q < 0.1; Supplementary Table 4). Events more frequent in CASP8-mutated tumors included +9p (64% versus 39.1%) and mutations in HRAS (36% versus 2.7%), FAT1 (52% versus 20.8%), EPHA2 (18% versus 2.4%), EP300 (18% versus 4.6%) and NOTCH1 (38% versus16.2%). TP53 (50% versus 82.2%) and NSD1 (4% versus 14%) mutations and losses −17p (18% versus 59.3%), −3p (30% versus 87.3%) and −11q (8% versus 46.4%) were significantly less prevalent. CASP8-mutated tumors thus are evidently driven more by mutations in multiple driver genes than by chromosome arm gains or losses. Single-individual timing diagrams of CASP8-mutated cases show early CASP8, TP53 and CDKN2A mutations (Fig. 3d). CASP8 mutation itself had an mRT of 0.34 (Fig. 3e), similar to that of NSD1 in the NSD1-mutated subset. Four events were significantly earlier in CASP8-mutated HPV– HNSCC than in other HPV– HNSCC (Fig. 3f), although of these, only +5p had a prevalence higher than 20%. Two losses, −9p and −15q, were significantly later in CASP8-mutated tumors.
NOTCH1-mutated tumors showed only few differences from other HPV– HNSCCs in event prevalence or timing, including a higher prevalence of mutations in CASP8 (24.1% versus 9.1%) and lower prevalence of +17p (19.0% versus 31.0%) and +8p (3.8% versus 17.3%; Supplementary Table 6). Only −5q showed different timing from other HPV– HNSCCs (mRT of 0.15 versus 0.45; Extended Data Fig. 2), although fewer events and individuals in this subtype might have limited the power of this analysis (Extended Data Fig. 2d).
Overall, timing analysis of distinct tumor subtypes can detect differential ordering of events independent of prevalence and associated subtype-specific orders with the unique biology of each subtype (Extended Data Fig. 3).
Extended Data Fig. 3. Summary of the developmental trajectories of the main subtypes of HNSCC.
Prevalence versus median Relative Timing (mRT) estimates for major events in subsets of HNSCC, from top to bottom: HPV+, CASP8-mutated HPV–, NSD1-mutated HPV–, all HPV–.
Genetic heterogeneity, aneuploidy and progression
High levels of genetic heterogeneity in a tumor are associated with worse outcomes in HNSCC48–51 and other types of cancer52. In HNSCC, studies have used the mutant allele tumor heterogeneity (MATH) score as a measure of genetic heterogeneity, defined as the median-normalized width of the distribution of mutant allele fractions (MAFs; Fig. 4a–c and Extended Data Fig. 4)48. Heterogeneity of MAF values could arise from mutation multiplicity differences within cells or differences in mutations among subclones. PhylogicNDT analysis allowed us to investigate relationships between MATH and other measures of genetic heterogeneity, evaluate the genomic source of high MATH scores and estimate how the MATH score changes during progression.
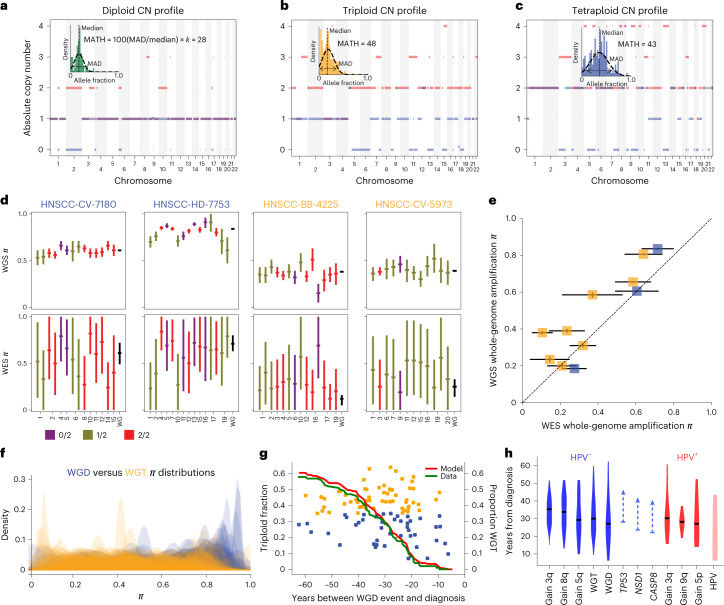
Fig. 4. Whole-genome events and aneuploidy classes in HNSCC.
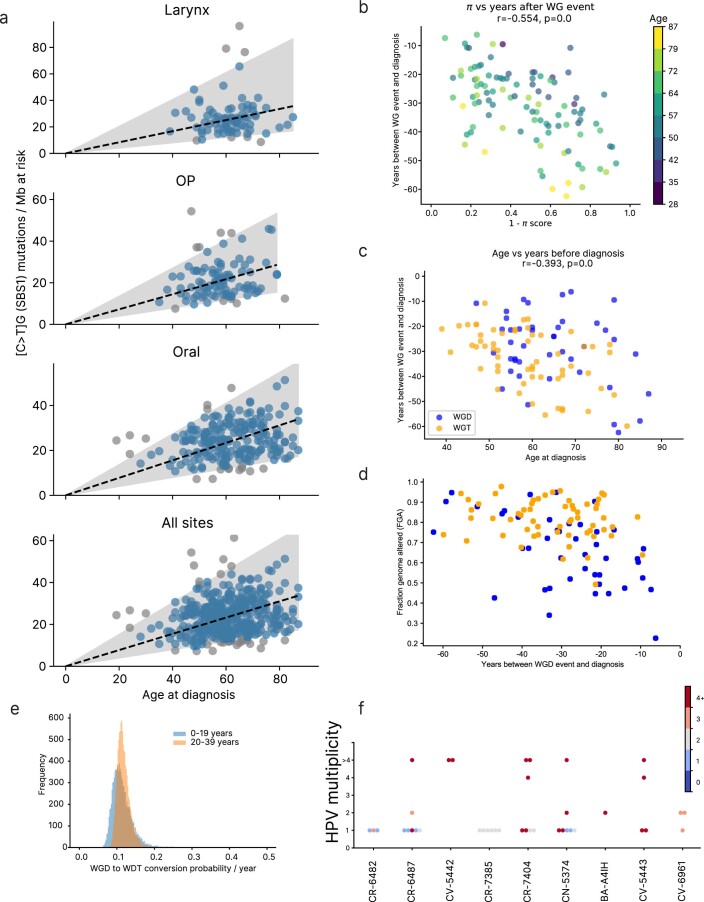
a–c, Allelic copy number (CN) profiles after ABSOLUTE in example tumors with diploid (a), triploid (b) and tetraploid (c) profiles. Each locus has a blue line and a red line representing the copy numbers of the minor and major alleles, respectively. Histograms of raw MAF distributions among somatically mutated loci and corresponding MATH values (scale factor of k = 1.4826 for the median absolute deviation (MAD) of a normal distribution to equal its standard deviation) are shown. d, Comparison of whole-genome amplification timing determined from WES against that from WGS of the same tumors (N = 4). Each chromosome arm is timed individually and aggregated to determine the timing of the whole-genome event. Left, two tumors with a tetraploid profile. Right, two tumors with a triploid profile. Horizontal ticks represent means. Error bars represent 75% credible intervals from posterior sampling. Colors represent copy number states. e, Scatter plot of whole-genome amplification timings in WGS versus WES of 11 tumors. Boxes represent timing means. Error bars represent 75% credible intervals from posterior sampling; blue, tetraploid profile; yellow, triploid profile. f, Timing probability distributions across tumors (N = 216 whole-genome amplifications), on π scale, of whole-genome amplifications leading to tetraploid (WGD) and triploid (WGT) copy number profiles. g, Real-time timing of WGT and WGD events. Estimated conversion rate of WGD events into WGT per year with a Poisson-like model (red model and green data; N = 103 real-timed whole-genome amplifications). h, Real-time timing of main driver events in HPV+ (red) and HPV– (blue) tumors (N = 205 individuals with real-timed driver events). Arrows for somatic point mutations represent that the estimate is late bound by the time of the regional gain, but no early bound exists. HPV integration sites include early events and events occurring during the development of the tumor.
Extended Data Fig. 4. Absolute copy number profiles.
Copy-number (CN) profiles from SNP arrays or whole-exome sequencing (WES; after ABSOLUTE correction for purity) of alleles in example tumors with diploid (a), triploid (b), and tetraploid (c) profiles. Each locus has a blue line and a red line representing the copy number of the minor and major alleles respectively.
We identified three classes of tumor aneuploidy based on absolute copy number profiles (Fig. 4). About half of tumors (Fig. 4a) showed essentially diploid profiles, with some LOH or copy number gains. Tumors with more disrupted copy number profiles, suggesting a genome-wide amplification during tumorigenesis, unexpectedly manifested as two distinct subtypes with similar prevalence: (1) triploid cancers with multiple genomic regions showing three total copies (WGT; Fig. 4b) and (2) tetraploid cancers with multiple genomic regions at four copies (two copies of each allele; Fig. 4c), suggesting a WGD event. Examples of single-tumor timing of these aneuploidy classes are in Extended Data Fig. 1.
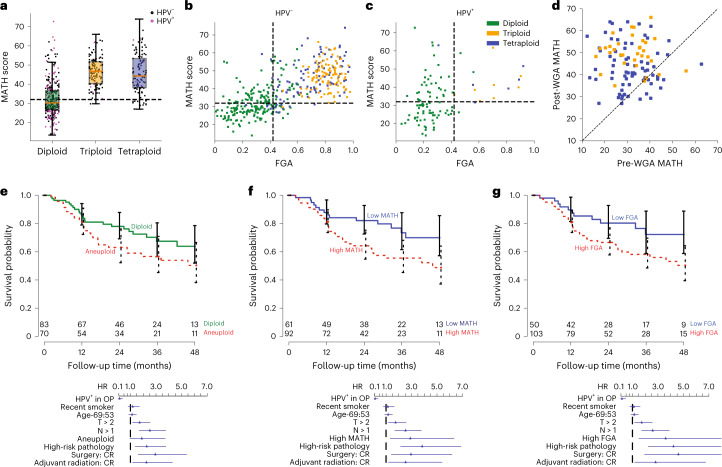
We investigated timing differences between WGT and WGD events. WGT occurred significantly earlier in both WES and whole-genome sequencing (WGS) single-tumor data (rank-sum test, P < 0.001; Fig. 4d–f), suggesting that primary tumors with WGT profiles experienced early WGD events, providing sufficient time to delete genomic regions that brings the average copy number down to approximately three. More recent genome duplication in tetraploid tumors (labeled WGD) kept the average copy number close to four. Increased aneuploidy going from diploid to WGD and WGT stages (Fig. 4a–c and Extended Data Figs. 4 and 5) and wider distributions of MAFs (higher MATH scores) with higher aneuploidy support this interpretation. Higher aneuploidy classes were associated with higher MATH values (Fig. 5a) and were thus related to that clinically relevant measure48–50. The fraction of the genome altered (FGA), reflecting the genomic regions with copy numbers different from the modal copy number level, provided an overall measure of aneuploidy. Noticeably, this measure was near linearly related to the MATH score (Fig. 5b) and strongly associated with aneuploidy class. The distribution of aneuploidy classes, FGA and MATH scores differed significantly between HPV+ and HPV– HNSCCs (Fig. 5b,c). The long-established relationship between HPV+ tumors and low MATH values48 is thus explained by lower prevalence of whole-genome events in HPV+ tumors.
Extended Data Fig. 5. Realtime timing.
(a) CpG mutation rate per bases at risk vs age of the patient. Robust linear regression with top and bottom 5% excluded (shaded area). (N = 413 patients) (b-d) Timing of WGT and WGD events in real time (b-e: N = 103 real-timed whole genome amplifications) (e) The posterior for Lambda in the Poisson process representing the conversion of WGD to WGT per year (f) HPV integration sites’ multiplicities for WGS data used in timing analysis (N = 52 HPV integration sites).
Fig. 5. Relationships among measures of intratumor genetic heterogeneity and their associations with outcome.
a, Box plots of MATH scores versus aneuploidy class for N = 522 total participants; red lines indicate the median, whiskers extend to the extreme values within 1.5× IQR of the box delimiting the first and third quartiles. b,c, Individual tumor MATH scores versus the FGA. Colors represent tumor aneuploidy classes. A Student’s t-test for correlation (bivariate data assumed but not tested) was conducted to obtain P values. Data are displayed for HPV– (b) and HPV+ (c) tumors. d, Timing relationship between MATH and WGD. For tumors with timing of WGD, MATH calculated from post-WGD SNV (vertical axis) is plotted against that calculated from pre-WGD SNV (horizontal axis). e–g, Associations of intratumor heterogeneity measures with outcome. Top, Kaplan–Meier survival curves (with 95% log survival CIs) stratified by a heterogeneity measure for the participant subset with survival most associated with intratumor heterogeneity (surgery without adjuvant therapy or therapy involving chemoradiation; excluding HPV+ oropharyngeal tumors to avoid confounding with HPV; N = 153 participants). Numbers at risk are shown over time. Bottom, HR point estimates (with 95% Wald CI) from Cox multiple regression model on 441 participants. The model was stratified by anatomic site and included interactions of the heterogeneity measure with therapy received and with high-risk pathology (evidence of close or positive surgical margins or extranodal tumor extension). HRs are displayed for HPV+ oropharyngeal tumors versus others (HPV+ in OP), smoked within 15 years of diagnosis (recent smoker), 75th and 25th percentiles of age (Age-69:53), T classification greater than 2 (T > 2), N classification greater than 1 (N > 1), presence of high-risk pathology as defined above (high-risk pathology) and surgery without adjuvant radiation and surgery with adjuvant radiation alone versus those receiving chemoradiotherapy (CR) as primary therapy or adjuvant to surgery (Surgery: CR and Adjuvant radiation: CR), with aneuploidy as the heterogeneity measure (e), with high MATH (MATH > 32.7) as the heterogeneity measure (f) and with FGA as the heterogeneity measure (g); high FGA cutoff is at the same percentile among tumors as for MATH.
As PhylogicNDT provided timing information for whole-genome events in single tumors, we could estimate MATH values before such events by only including mutations timed before a tumor’s whole-genome event (Fig. 5d). MATH values were generally much higher after WGD, presumably due to marked aneuploidy after doubling. Although high MATH is found in many diploid tumors (Fig. 5b,c), it is almost universal (213/233, 91.4%) in tumors with a whole-genome event.
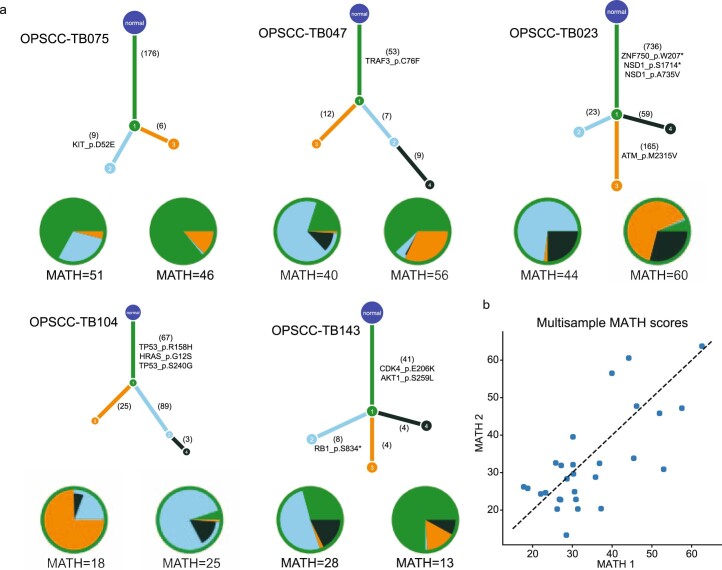
Paired samples (two from the same tumor) were available for 28 oropharyngeal tumors newly collected for this study. Although the mutations and subclonal composition could differ substantially between paired samples (Extended Data Fig. 6a), their overall MATH and FGA measures of heterogeneity were similar (Extended Data Fig. 6b). These measures seem to be intrinsic characteristics of a tumor as a whole, supporting their clinical potential as biomarkers.
Extended Data Fig. 6. Multi-sample patient phylogeny and heterogeneity.
(a) Examples of phylogenetic trees generated by PhylogicNDT (BuildTree module) for selected patients with two samples of the same tumor. Numbers on branches represent the number of SNVs or indels that are added along each branch. Pie graphs below represent cancer cell populations of each subclone, constrained by the pigeonhole principle, estimated by PhylogicNDT (BuildTree module). (b) Scatter plot of MATH scores from patients with multiple tumor samples. Each axis represents the MATH score from one of the samples. (N = 28 patients with multiple samples).
Time between somatic events, including HPV integration, and diagnosis
We converted relative timing estimates to the expected number of years before diagnosis. We modeled CpG>T (‘aging’ signature SBS1) mutations (total number of mutations/number of covered CpG sites) as a function of participant age at diagnosis (Extended Data Fig. 5a). We observed an accumulation of 0.37 (interquartile range (IQR) of 0.29–0.51) CpG>T mutations per megabase at risk per year in HNSCC HPV– tumors and a similar value of 0.39 (IQR of 0.25–0.45) in HPV+ tumors. We found that these rates were also similar among tumor anatomical sites (Extended Data Fig. 5a). We then used these rates to convert participant-specific CpG>T π estimates for driver events to real time, measured in years before diagnosis (Extended Data Fig. 5a–d).
Real-time timing estimates of WGT and WGD support a model with an abrupt transition from the WGD state to WGT (Fig. 4g, Extended Data Fig. 5c–e and Supplementary Table 7). Only two triploid tumors had the WGD event estimated to occur during the 15-year period before diagnosis, whereas seven WGD cases did not transition to WGT in that time frame (Fisher’s exact test P = 0.033 compared to WGT/WGD greater than 15 years before diagnosis).
A Poisson-like process model with a fixed rate of conversion from WGD to WGT per year (allowing for a fraction of WGD cases to never transform to WGT) fits the data well (Fig. 4g, red and green curves) and yields a relatively narrow estimate for the conversion rate of 11 ± 2% per year (Fig. 4g and Extended Data Fig. 5c–e). About 35% of tumors never reached triploidy even if the WGD event happened decades before diagnosis.
The relationship between FGA and the timing of the WGD/WGT event (Extended Data Fig. 5d) further supported an abrupt WGD-to-WGT transition. In a gradual process, cases with earlier WGD would slowly increase FGA over time. We instead found only a minor correlation in the years closest to diagnosis.
For other major somatic events, the earliest events in HPV– cancers (+3q, +8q and TP53) occur around 30–40 years before diagnosis, while HPV+ early events (+3q and +9q) occur around 20–30 years before diagnosis (Fig. 4h). More recent clonal events, occurring less than 15–20 years before diagnosis, can be associated with clonal expansion, leading to primary disease. Although WGD events can occur very early, most occur ~20–25 years before diagnosis. All WGT tumors had the event 10 or more years before diagnosis. These time scales are consistent with the slow conversion rates from dysplasia to invasive carcinoma, on the order of 1 to 4% per year, reported for HPV– HNSCC53.
Additionally, we estimated the timing of HPV integration events. We first aligned reads to the structural variation breakpoints at boundaries of 53 HPV integration sites across 11 individuals with WGS data (HPV can integrate multiple times per individual39) and analyzed multiplicities and CCFs (Extended Data Fig. 5f); 40 sites in 9 individuals showed good coverage. Many integration sites had high multiplicity on early gains, suggesting that integration of HPV is a pregain event contributing to initiating tumorigenesis. We also identified some lower-multiplicity and subclonal integration sites, suggesting that HPV integration is a continuous process that does not stop during progression (Fig. 4h and Extended Data Fig. 5f). Converting the timing of HPV+ integration into years before diagnosis suggests that initial HPV integration events can happen more than 25 years before diagnosis, with additional integrations throughout tumor development (Fig. 4h).
In summary, our timing analysis shows that (1) tumors are typically diagnosed many years after key driver events, which can happen at different ages (Extended Data Fig. 5), (2) founding events can occur 25–35 years or more before diagnosis, (3) WGT events do not seem to occur in the last ~10 years before diagnosis, and (4) HPV integration sites occur early in tumor development so that HPV, as expected, is the main contributor to HPV+ cancer initiation.
Associations of heterogeneity and aneuploidy with outcome
Associations of HPV– status and high MATH score with shorter overall survival in HNSCC are well established49–52. To see whether FGA and whole-genome events are similarly associated with outcome, we first examined a large subset of individuals whose survival is most strongly associated with intratumor heterogeneity: those without high-risk pathologic features who received either chemoradiation therapy or surgery without adjuvant therapy51. Initial analysis was restricted to individuals with HPV– tumors to remove confounding associations. Among these 145 individuals (with 53 deaths), high FGA showed an association with overall survival similar to that of high MATH. High- versus low-score hazard ratios (HRs; with 95% confidence interval (CI95%)) were FGA (2.09; 1.05–4.2) and MATH (2.08; 1.11–3.9). Aneuploid tumors or tumors with whole-genome events leading to WGD or WGT had weaker associations with overall survival (aneuploid/diploid HR of 1.73 and CI95% of 0.99–3.0; whole-genome event/none HR of 1.54 and CI95% of 0.89–2.7; Fig. 5e–g).
To evaluate these associations in more individuals, including those with HPV+ tumors, while taking other outcome-associated variables into account, we extended our published survival model with MATH51 to incorporate the additional cases in this study. MATH was significantly associated overall with outcome (chi-square 17.75, 4 d.f., P = 0.0014, Wald test). Replacing MATH with FGA or WGD/WGT (Fig. 5e–g) in an otherwise identical model also showed significant associations with outcome (FGA: chi-square 12.68, 4 d.f., P = 0.013; WGD/WGT: chi-square 10.32, 4 d.f., P = 0.035). Aneuploidy class analyzed similarly was just above the significance threshold (chi-square 8.97, P = 0.06). Applying the Akaike information criterion (AIC), MATH (AIC of 1,350) had the strongest association with survival, although genomic instability measures had similar AIC scores (FGA, 1,355; WGD/WGT, 1,358; aneuploidy, 1,360).
Early genetic progression and aneuploidy development in tumors thus can be modeled computationally using WES data from primary tumors, providing findings about processes strongly associated with survival. Timing analysis is possible even when premalignant tissue is unobtainable, as in HPV+ HNSCC.
Discussion
Our analysis of HPV+ HNSCC fills a major gap in knowledge about the progression of genetic events in this increasingly prevalent and clinically significant disease35,37, a cancer whose lack of premalignant tissue frustrated prior attempts at timing39,40. Analysis of HPV– HNSCC verified that our computational approach could recapitulate the progression in classic HPV– disease8,54 and extend the progression model to additional events in significant subsets of HPV– disease. These results on cohorts of primary HNSCC specimens suggest that this approach could be applied to other types of cancer whose early genetic progression is unknown because premalignant tissue is difficult or impossible to obtain.
We extended the genetic progression model for HPV– HNSCC to a total of 61 SNV or copy number alteration events, of which 17 had >40% prevalence. Our timing estimates agreed well with the Califano et al. model8 and with more recent studies that refined and extended it30–34. Comparing predicted mRT values (Fig. 1e) for events associated with the progression stages reported by Califano et al.8 (Fig. 1a) allowed us to map pathologic disease stages onto the mRT timeline. We determined when whole-genome events leading to WGD or WGT occur during HPV– tumorigenesis, yielding estimates before or near the clinical development of invasive cancer.
Additionally, we identified genes with early mutations that likely play roles in HPV– tumor initiation (FBXW7, NOTCH1, CASP8, FAT1, NSD1, HRAS, EP300, CREBBP and KMT2D), with over 60% of HPV– HNSCCs having a mutation in at least one of these genes. Of those, we found anatomic and genetic differences among tumors, with CASP8 mutations almost solely in the oral cavity and NSD1 predominant in laryngeal tumors. By contrast, tumors with NOTCH1 mutations, long suspected to play a role in HPV– HNSCC, showed no anatomic preference or frequency or timing differences from other HPV– HNSCC. Genes with mutations at later mRT (PK3CA, NFE2L2, HLA-A, SPEN and KMT2C) are more likely to be important for progressing to later stages of the disease.
In HPV+ HNSCC, gain of chromosome arm 3q and loss of arm 11q are both earlier and more frequent than in HPV– HNSCC, indicating important early roles during HPV+ progression. We estimate that genomic integration of HPV viral DNA occurs early in tumor development, consistent with HPV being the main contributor to cancer initiation.
Arm 3q includes the PIK3CA locus, whose activation by copy number gain or mutation occurs often in HNSCC43,55,56; 74% of HPV+ and 62% of HPV– HNSCC showed genetic alterations related to the PIK3CA locus. Gain of 3q was found at an earlier mRT in HPV+ tumors (0.05) than in HPV– tumors (0.51; Figs. 1e and 2d,e), suggesting a particularly important early role for amplification of PIK3CA in HPV+ disease. Notably, however, mutations in PIK3CA, targeted by some therapies57, were relatively late in genetic progression in both classes of HNSCC. If tumors are less ‘addicted’58 to mutations that occur late in tumor development, therapies targeted against PIK3CA mutations might be of limited effectiveness.
With respect to loss of 11q in HPV+ HNSCC, ATM stands out as a candidate for early involvement in tumorigenesis. Although ATM improves the ability of episomal HPV to replicate59,60, its role in protection against double-stranded DNA breaks60,61 would be expected to inhibit genomic integration of HPV DNA. Losses of 11q also occur in HPV– disease but much later at an mRT of 0.67 that we estimate to be near the development of carcinoma in situ (Figs. 1e and Fig. 2d,e).
Identifying genes whose loss along with chromosome arm 3p promote development of HPV– HNSCC has long been an active area of interest62. The surprisingly early timing and high prevalence of 3p loss that we also found in HPV+ HNSCC suggests that studies of 3p genes will have clinical significance for all HNSCC.
Similarly surprising in HPV+ HNSCC is the prevalence of 17p loss, which includes the TP53 locus, occurring in one-quarter of tumors (26 of 101). As HPV E6 leads to inactivation of the p53 protein product42, and TP53 mutations occur in only 3% of HPV+ HNSCC, loss of additional genes on 17p presumably are important. Rather than the very early loss seen in HPV– tumors, 17p loss occurs at intermediate stages of HPV+ disease progression (mRT of 0.66), suggesting that those genes are involved in later stages of tumor development.
Using a clock-like mutational signature to convert relative event timing estimates to years before diagnosis, we identified that early founding events can occur as many as 30–40 years before tumor diagnosis in HPV– disease and 20–30 or more years in HPV+ tumors, with HPV integration also early in the development. WGD-to-WGT conversion seems to follow an abrupt transition model, with a specific transition probability per year.
This work helps clarify the nature of outcome-associated genomic disruption measures. Almost all tumors with a whole-genome event leading to WGT or WGD had high MATH values. Similar to high MATH, high FGA was also associated with shorter overall survival in HNSCC when therapies and standard clinical and pathologic characteristics were taken into account. Measures of genomic disruption can be used to classify individuals into high- and low-risk groups for monitoring disease with strategies such as minimal residual disease assays63. Identification of WGD timing before invasion might be used to identify HPV– HNSCC at higher risk of progression. Other early drivers can be the basis of additional therapeutic trials, including in the preventive setting for high-risk individuals.
Applying our approach to other cancers whose premalignant tissue is seldom or never obtainable14–22, including multiple rare tumor types, could be of great clinical importance. Many such cancers have uncertain etiology, with effective treatment avenues less well defined than in more prevalent tumors. Establishment of progression trajectories, early drivers and similarity of progression to other cancer types can lead to development of improved disease models, better treatment strategies and clinical management and more accurate prediction of disease course.
Methods
This research complied with all relevant ethical regulations. Human research on individuals at Massachusetts Eye and Ear (MEE) was approved by the MEE Human Studies Committee under protocol HSC 11-024H, with informed consent obtained from participants. Participants were not compensated.
Participants, clinical data and tumors
We analyzed data from 486 individuals with head and neck cancer in TCGA43 and 45 individuals from MEE with oropharyngeal tumors whose WES data met quality control criteria (see below). TCGA clinical data were as in previous work50,51. Clinical data from MEE corresponding to TCGA data fields were extracted from participant records. Median time to death for 218 participants was 13.7 months (IQR of 8.4 to 26.5 months); follow-up time for the 313 last known to be alive was approximately twice as long (median of 29.4 months; IQR of 17.1 to 51.2 months). HPV status of TCGA tumors was assessed by RNA sequencing43; for MEE tumors, clinical HPV annotations were used. Clinical data are provided as Source Data for Fig. 5, including information on age and sex. Survival analysis was limited to individuals who survived more than 60 d after definitive pathologic diagnosis so that associations of outcome with adjuvant therapy could be ascertained.
TCGA tumor and control tissue or blood samples were as previously reported43. Tumor portions from MEE participants were frozen at liquid nitrogen temperature; participant-matched frozen blood provided control DNA for assessing tumor-specific mutations. Twenty-eight of the MEE tumor samples had two subsamples taken for separate processing.
WES
Tumors from TCGA had undergone WES or WGS, as described25. Analysis proceeded from paired aligned bam files, which were inputted into a standard WES somatic variant-calling pipeline, including MuTect for calling somatic SNVs64, Strelka for calling small insertions and deletions65, deTiN for estimating tumor-in-normal contamination66, ContEst for estimating cross-participant contamination67, AllelicCapSeg for calling allelic copy number variants68 and ABSOLUTE for estimating tumor purity, ploidy, CCFs and absolute allelic copy number68. Artifactual variants were filtered out using a token panel of normals filter, a blat filter and an oxoG filter. For TCGA tumors in which the WES data did not yield sufficiently high-quality copy number data from AllelicCapSeg (361 tumors), we used HapSeg on single-nucleotide polymorphism (SNP) arrays as a substitute step. MEE tumors underwent a similar variant-calling pipeline as the TCGA tumors, but we did not substitute any AllelicCapSeg results with HapSeg on SNP arrays.
Signature analysis for CoMut plots
We performed SBS signature analysis separately on the HPV– and HPV+ subsets using SignatureAnalyzer69,70 on the somatic SNV calls resulting from the variant-calling pipeline.
PhylogicNDT
We used PhylogicNDT to estimate relative event timing within individual tumors (SinglePatientTiming) and to combine timing information among tumors (LeagueModel). This analysis suite has been described in detail elsewhere26.
Within-tumor timing
After ABSOLUTE analysis68 to determine purity and ploidy, allele-specific SNV multiplicity estimates and purity-corrected copy number variation values from WES or WGS reads were used to set a within-tumor partial ordering of events. For example, on a chromosomal region that has been doubled, a tumor-specific mutation present at two copies is timed before the doubling, while a mutation present at only one copy is timed after the doubling. Tumor-specific genetic events are mapped on a π scale (as a probability distribution) from 0 to 1 (refs. 23,26), representing the first and last clonal genetic events. Estimated distributions are corrected for power to detect based on coverage profiles. The proportion of clonal events per megabase occurring before each genetic event in a tumor is defined as the π score for that event. Clonal events that cannot be timed are assigned a uniform π distribution, and the last clonal and all subclonal events are assigned π scores of 1, represented as a delta function δ(π − 1). Uncertainties arising from sequencing are incorporated into a distribution of π values for each event within the tumor. Thus within-tumor timing uses information from all tumor-specific SNV and copy number variation events.
Across-tumor timing
PhylogicNDT LeagueModel uses a Bayesian Monte Carlo Markov chain (MCMC) approach to combine single-tumor π-score distributions for each genetic event shared among a number of tumors into a consensus genetic relative timing progression. This consensus relative timing is generated by repeated sampling from the single-tumor π distributions of the shared events among multiple subsets of tumors. On this relative timing scale, a value of 0 represents the earliest shared event, and 1 is the last clonal shared event. The result is a distribution of relative timing scores for each shared event among the tumors based on comparison of within-tumor π scores among shared events. This was repeated for 200 iterations via resampling at an average of 63% without replacement, and the union of the relative timing score traces for each iteration was used as the final timing score distribution.
Timing comparisons
To compare consensus genetic progressions between HPV+ and HPV– HNSCC or between subsets of HPV– tumors, we performed a similar MCMC approach to the LeagueModel algorithm, but for each resampling iteration, we calculated the difference in the relative timing scores between subsets. Point estimates for timing differences were calculated as the median of the final trace, and one-way P values were calculated as the proportion of trace samples that was greater than 0. These were converted to two-way P values (P2-way = 1 − 2|0.5 − P1-way|) and then to q values using multiple hypothesis correction.
Real-time timing of somatic events
To estimate the real-time timing of somatic events, we used a robust linear regression model by removing the top and bottom 5% of slope outliers to fit the rate of clonal CpG>T (the ‘aging’ signature, SBS1) mutations (total number of mutations/number of covered CpG sites) corrected for copy number and multiplicity as a function of the age at diagnosis (Extended Data Fig. 5a) across different anatomical sites. The CpG>T rate per year and participant age were used to calculate the posterior distribution of the individual somatic event real-time timing measured from the π-score distribution calculated by using only CpG>T mutations with PhylogicNDT SinglePatientTiming. The resulting distribution for a specific event was then combined across participants to produce a cohort-level distribution for the typical time of the event in terms of years before diagnosis. Timing of HPV integration events was performed by aligning structural variation breakpoints and spanning reads at 53 integration sites across 11 individuals with available WGS data and performing a similar timing analysis as for mutations (using local copy number and clonal multiplicity). Because APOBEC mutations are often clustered near HPV integration sites, we excluded APOBEC mutations for the timing estimate. Often several integration sites were identified per participant, and sites on copy number regions with the earliest estimate of real time were used as representative for the participant.
Measures of intratumor genetic heterogeneity
MATH was calculated per tumor sample as previously described48. For tumors represented by two separately sequenced samples, MATH values were averaged.
A tumor’s ploidy group was assigned based on the fraction of the genome in diploid, triploid and tetraploid states. Tumors with a diploid fraction of >0.65 were classified as diploid, non-diploid tumors with a fraction triploid of >0.35 were classified as triploid, and other non-diploid tumors were classified as tetraploid.
A whole-genome event was called if at least half of chromosome arms were amplified or at least four chromosome arms were amplified on both alleles. Further classification into triploidy (WGT) or doubling (WGD) was based on the corresponding ploidy group (triploid or tetraploid).
The FGA absent a whole-genome event was the fraction of the genome deviating by more than 0.2 from a value of one copy for either allele. With a whole-genome event, FGA was calculated as the fraction of genome deviating by more than 0.2 from a value of two copies for either allele.
Statistical analysis and reproducibility
No statistical method was used to predetermine sample size. As there were no groups defined by experimental manipulations, no blinding was performed. Random resampling is inherent in PhylogicNDT, as described above.
Event prevalence comparisons, associations among measures of intratumor heterogeneity and other routine analyses were performed with standard statistical routines in Python or R. Frequentist tests (except for inherently one-sided chi-square statistics) were two sided. The Benjamini–Hochberg false discovery rate correction was applied to event prevalence comparison P values within each HNSCC subset. Survival analysis used the survival71 (version 3.3-1) and rms72 (version 6.3-0) packages under version 4.2.0 of R73, extending a previously described model for HNSCC survival51. Cox survival models were stratified by anatomic site to satisfy the proportional hazards assumption, and model calibration was checked by bootstrap resampling.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Supplementary Table 1. Event prevalence comparison between HPV+ and HPV– HNSCC. a, Number and percentage of cases having each of 66 genomic events for 421 HPV– and 101 HPV+ HNSCCs. Fisher’s exact test two-sided P values are reported for comparison between HPV– and HPV+ tumors. Benjamini–Hochberg false discovery rate-corrected q values are also shown. b, List of differentially prevalent events at q < 0.1. c, List of events not differentially prevalent (q < 0.1). d, Differentially prevalent events in b favored in HPV– HNSCC. e, Differentially prevalent events in b favored in HPV+ HNSCC. Supplementary Table 2. Clonal event prevalence comparison between HPV+ and HPV– HNSCC, like Supplementary Table 1 but restricted to clonal events. Supplementary Table 3. Event prevalence by anatomic subsite for HPV– HNSCC. Breakdown by anatomic subsite of number and percentage of cases having specified genomic events: larynx, 114 tumors; oral cavity, 274 tumors; oropharynx, 33 tumors. Fisher’s exact test P values and Benjamini–Hochberg q values are as in Supplementary Table 1. Supplementary Table 4. Event prevalence comparison between NSD1-mutated and other HPV– HNSCC, like Supplementary Table 1a–c but comparing 54 NSD1-mutated HPV– HNSCC against 367 other HPV– HNSCC. Supplementary Table 5. Event prevalence comparison between CASP8-mutated and other HPV– HNSCC, like Supplementary Table 1a–c but comparing 50 CASP8-mutated HPV– HNSCCs against 371 other HPV– HNSCCs. Supplementary Table 6. Event prevalence comparison between NOTCH1-mutated and other HPV– HNSCCs, like Supplementary Table 1a–c but comparing 70 NOTCH1-mutated HPV– HNSCCs against 342 other HPV– HNSCCs. Supplementary Table 7. Real-time timing estimates for whole-genome copy-number events.
Acknowledgements
G.G., I.L., E.A.M. and J.W.R. were partially funded by the NIDCR (R01DE022087, to J.W.R.). G.G. is partially funded by the Paul C. Zamecnick, MD, Chair in Oncology at Massachusetts General Hospital. G.G. and I.L. were partially funded by grant U2CCA233238 from the National Cancer Institute. J.W.R. is partially funded by the Mary E. and John W. Alford Chair and The Joan Bisesi Fund for Head and Neck Cancer Research at The Ohio State University Wexner Medical Center and the Ohio State University Comprehensive Cancer Center. Additional funds were provided by the Ohio State University Comprehensive Cancer Center, which is supported by grant P30CA016058 from the National Cancer Institute.
Extended data
Source data
Source data for Figs. 1–5 and Extended Data Figs. 2 and 4–6.
Author contributions
I.L., E.A.M., G.G. and J.W.R. developed the idea for the study. D.T.L. and J.W.R. collected tumor and blood specimens from consented participants at MEE, which were further processed by E.A.M., J.B.-V., A.L.-T. and W.A.M. W.C.F. was responsible for pathologic evaluation of tumors from MEE participants, including HPV status. Clinical data from MEE were collected and processed by J.B.-V., A.L.-T. and E.A.M. I.L., J.C., D.R. and O.S. performed initial genomic analysis, timing analysis and comparisons of timing between subsets of HNSCC. I.L. and J.C. performed all other genomic analyses and correlations with heterogeneity measurements. All authors made intellectual contributions over the course of the study. I.L., E.A.M., J.C., G.G. and J.W.R. were responsible for preparing the manuscript.
Peer review
Peer review information
Nature Cancer thanks David Sidransky, Moritz Gerstung and Luc Morris for their contribution to the peer review of this work.
Data availability
New WES data that support the findings of this study have been deposited in dbGaP under accession code phs003139.v1. Other human HNSCC genomic data were derived from the TCGA Research Network at http://cancergenome.nih.gov/. Source data are provided with this paper. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
Code availability
The PhylogicNDT package is available for download from https://github.com/broadinstitute/PhylogicNDT.
Competing interests
G.G. receives research funds from IBM and Pharmacyclics and is an inventor on patent applications related to MSMuTect, MSMutSig, MSIDetect, POLYSOLVER and TensorQTL. G.G. is a founder of, consultant for and holds privately held equity in Scorpion Therapeutics. I.L. is a consultant for PACT Pharma, Inc., and a Board member, scientific adviser and consultant and holds privately held equity in ennov1, LLC and NoRD Bio, Inc. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ignaty Leshchiner, Edmund A. Mroz, Justin Cha.
These authors jointly supervised this work: Gad Getz, James W. Rocco.
Contributor Information
Gad Getz, Email: gadgetz@broadinstitute.org.
James W. Rocco, Email: james.rocco@osumc.edu
Extended data
is available for this paper at 10.1038/s43018-023-00533-y.
Supplementary information
The online version contains supplementary material available at 10.1038/s43018-023-00533-y.
References
- 1.Garnis C, Buys TP, Lam WL. Genetic alteration and gene expression modulation during cancer progression. Mol. Cancer. 2004;3:9. doi: 10.1186/1476-4598-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Srivastava S, Grizzle WE. Biomarkers and the genetics of early neoplastic lesions. Cancer Biomark. 2011;9:41–64. doi: 10.3233/CBM-2011-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sen S, Hopwood V. Molecular cytogenetic evidence for multistep tumorigenesis: implications for risk assessment and early detection. Cancer Biomark. 2011;9:113–132. doi: 10.3233/CBM-2011-0171. [DOI] [PubMed] [Google Scholar]
- 4.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- 5.Manne U, Shanmugam C, Katkoori VR, Bumpers HL, Grizzle WE. Development and progression of colorectal neoplasia. Cancer Biomark. 2011;9:235–265. doi: 10.3233/CBM-2011-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiltshire RN, et al. Direct visualization of the clonal progression of primary cutaneous melanoma: application of tissue microdissection and comparative genomic hybridization. Cancer Res. 1995;55:3954–3957. [PubMed] [Google Scholar]
- 7.Bastian BC, LeBoit PE, Hamm H, Bröcker EB, Pinkel D. Chromosomal gains and losses in primary cutaneous melanomas detected by comparative genomic hybridization. Cancer Res. 1998;58:2170–2175. [PubMed] [Google Scholar]
- 8.Califano J, et al. Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res. 1996;56:2488–2492. [PubMed] [Google Scholar]
- 9.Park BJ, Chiosea SI, Grandis JR. Molecular changes in the multistage pathogenesis of head and neck cancer. Cancer Biomark. 2011;9:325–339. doi: 10.3233/CBM-2011-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reid BJ. Early events during neoplastic progression in Barrett’s esophagus. Cancer Biomark. 2011;9:307–324. doi: 10.3233/CBM-2011-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurmyshkina OV, Kovchur PI, Volkova TO. ‘Drawing’ a molecular portrait of CIN and cervical cancer: a review of genome-wide molecular profiling data. Asian Pac. J. Cancer Prev. 2015;16:4477–4487. doi: 10.7314/APJCP.2015.16.11.4477. [DOI] [PubMed] [Google Scholar]
- 12.Czerniak B. Molecular pathology and biomarkers of bladder cancer. Cancer Biomark. 2011;9:159–176. doi: 10.3233/CBM-2011-0175. [DOI] [PubMed] [Google Scholar]
- 13.Cazares LH, et al. Molecular pathology of prostate cancer. Cancer Biomark. 2011;9:441–459. doi: 10.3233/CBM-2011-0181. [DOI] [PubMed] [Google Scholar]
- 14.Adamson DC, Rasheed BAK, McLendon RE, Bigner DD. Central nervous system. Cancer Biomark. 2011;9:193–210. doi: 10.3233/CBM-2011-0177. [DOI] [PubMed] [Google Scholar]
- 15.Block T, Mehta AS, London WT. Hepatocellular carcinoma of the liver. Cancer Biomark. 2011;9:375–383. doi: 10.3233/CBM-2011-0165. [DOI] [PubMed] [Google Scholar]
- 16.Cairns P. Renal cell carcinoma. Cancer Biomark. 2011;9:461–473. doi: 10.3233/CBM-2011-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole K, Tabernero M, Anderson KS. Biologic characteristics of premalignant breast disease. Cancer Biomark. 2011;9:177–192. doi: 10.3233/CBM-2011-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.David S, Meltzer SJ. Stomach—genetic and epigenetic alterations of preneoplastic and neoplastic lesions. Cancer Biomark. 2011;9:493–507. doi: 10.3233/CBM-2011-0169. [DOI] [PubMed] [Google Scholar]
- 19.Gazdar AF, Brambilla E. Preneoplasia of lung cancer. Cancer Biomark. 2011;9:385–396. doi: 10.3233/CBM-2011-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merritt MA, Cramer DW. Molecular pathogenesis of endometrial and ovarian cancer. Cancer Biomark. 2011;9:287–305. doi: 10.3233/CBM-2011-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Powers M, Zhang W, Lopez-Terrada D, Czerniak BA, Lazar AJ. The molecular pathology of sarcomas. Cancer Biomark. 2011;9:475–491. doi: 10.3233/CBM-2011-0170. [DOI] [PubMed] [Google Scholar]
- 22.Remmers N, Bailey JM, Mohr AM, Hollingsworth MA. Molecular pathology of early pancreatic cancer. Cancer Biomark. 2011;9:421–440. doi: 10.3233/CBM-2011-0168. [DOI] [PubMed] [Google Scholar]
- 23.Purdom E, et al. Methods and challenges in timing chromosomal abnormalities within cancer samples. Bioinformatics. 2013;29:3113–3120. doi: 10.1093/bioinformatics/btt546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nik-Zainal S, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gruber M, et al. Growth dynamics in naturally progressing chronic lymphocytic leukaemia. Nature. 2019;570:474–479. doi: 10.1038/s41586-019-1252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leshchiner, I. et al. Comprehensive analysis of tumour initiation, spatial and temporal progression under multiple lines of treatment. Preprint at bioRxiv10.1101/508127 (2019).
- 27.Parikh AR, et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 2019;25:1415–1421. doi: 10.1038/s41591-019-0561-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerstung M, et al. The evolutionary history of 2,658 cancers. Nature. 2020;578:122–128. doi: 10.1038/s41586-019-1907-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dentro SC, et al. Characterizing genetic intra-tumor heterogeneity across 2,658 human cancer genomes. Cell. 2021;184:2239–2254. doi: 10.1016/j.cell.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Houten VM, et al. Mutated p53 as a molecular marker for the diagnosis of head and neck cancer. J. Pathol. 2002;198:476–486. doi: 10.1002/path.1242. [DOI] [PubMed] [Google Scholar]
- 31.Tabor MP, et al. Comparative molecular and histological grading of epithelial dysplasia of the oral cavity and the oropharynx. J. Pathol. 2003;199:354–360. doi: 10.1002/path.1285. [DOI] [PubMed] [Google Scholar]
- 32.Ghosh A, et al. SH3GL2 and CDKN2A/2B loci are independently altered in early dysplastic lesions of head and neck: correlation with HPV infection and tobacco habit. J. Pathol. 2009;217:408–419. doi: 10.1002/path.2464. [DOI] [PubMed] [Google Scholar]
- 33.Bhattacharya A, et al. Two distinct routes to oral cancer differing in genome instability and risk for cervical node metastasis. Clin. Cancer Res. 2011;17:7024–7034. doi: 10.1158/1078-0432.CCR-11-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veeramachaneni R, et al. Analysis of head and neck carcinoma progression reveals novel and relevant stage-specific changes associated with immortalisation and malignancy. Sci Rep. 2019;9:11992. doi: 10.1038/s41598-019-48229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson DE, et al. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers. 2020;6:92. doi: 10.1038/s41572-020-00224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wijetunga NA, Yu Y, Morris LG, Lee N, Riaz N. The head and neck cancer genome in the era of immunotherapy. Oral Oncol. 2021;112:105040. doi: 10.1016/j.oraloncology.2020.105040. [DOI] [PubMed] [Google Scholar]
- 37.Chaturvedi AK, Engels EA, Anderson WF, Gillison ML. Incidence trends for human papillomavirus-related and -unrelated oral squamous cell carcinomas in the United States. J. Clin. Oncol. 2008;26:612–619. doi: 10.1200/JCO.2007.14.1713. [DOI] [PubMed] [Google Scholar]
- 38.Gillison ML, Chaturvedi AK, Anderson WF, Fakhry C. Epidemiology of human papillomavirus-positive head and neck squamous cell carcinoma. J. Clin. Oncol. 2015;33:3235–3242. doi: 10.1200/JCO.2015.61.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gillison ML, et al. Human papillomavirus and the landscape of secondary genetic alterations in oral cancers. Genome Res. 2019;29:1–17. doi: 10.1101/gr.241141.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chae J, et al. Genomic characterization of clonal evolution during oropharyngeal carcinogenesis driven by human papillomavirus 16. BMB Rep. 2018;51:584–589. doi: 10.5483/BMBRep.2018.51.11.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munger K, Jones DL. Human papillomavirus carcinogenesis: an identity crisis in the retinoblastoma tumor suppressor pathway. J. Virol. 2015;89:4708–4711. doi: 10.1128/JVI.03486-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wallace NA, Galloway DA. Novel functions of the human papillomavirus E6 oncoproteins. Annu. Rev. Virol. 2015;2:403–423. doi: 10.1146/annurev-virology-100114-055021. [DOI] [PubMed] [Google Scholar]
- 43.Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Black CC, Ogomo C. Does pTis exist in HPV-driven tonsillar carcinomas? An ultrastructural review and examination of two cases. Ultrastruct. Pathol. 2017;41:55–61. doi: 10.1080/01913123.2016.1258020. [DOI] [PubMed] [Google Scholar]
- 45.Perry ME, Jones MM, Mustafa Y. Structure of the crypt epithelium in human palatine tonsils. Acta Otolaryngol. Suppl. 1988;454:53–59. doi: 10.3109/00016488809125005. [DOI] [PubMed] [Google Scholar]
- 46.Perry ME. The specialised structure of crypt epithelium in the human palatine tonsil and its functional significance. J. Anat. 1994;185:111–127. [PMC free article] [PubMed] [Google Scholar]
- 47.Boscolo-Rizzo P, Schroeder L, Romeo S, Pawlita M. The prevalence of human papillomavirus in squamous cell carcinoma of unknown primary site metastatic to neck lymph nodes: a systematic review. Clin. Exp. Metastasis. 2015;32:835–845. doi: 10.1007/s10585-015-9744-z. [DOI] [PubMed] [Google Scholar]
- 48.Mroz EA, Rocco JW. MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol. 2013;49:211–215. doi: 10.1016/j.oraloncology.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mroz EA, et al. High intratumor genetic heterogeneity is related to worse outcome in patients with head and neck squamous cell carcinoma. Cancer. 2013;119:3034–3042. doi: 10.1002/cncr.28150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mroz EA, Tward AD, Hammon RJ, Ren Y, Rocco JW. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med. 2015;12:e1001786. doi: 10.1371/journal.pmed.1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mroz EA, Patel KB, Rocco JW. Intratumor heterogeneity could inform the use and type of postoperative adjuvant therapy in patients with head and neck squamous cell carcinoma. Cancer. 2020;126:1895–1904. doi: 10.1002/cncr.32742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu, T. et al. Intratumor heterogeneity as a prognostic factor in solid tumors: a systematic review and meta-analysis. Front. Oncol.11, 744064 (2021). [DOI] [PMC free article] [PubMed]
- 53.Iocca O, et al. Potentially malignant disorders of the oral cavity and oral dysplasia: a systematic review and meta-analysis of malignant transformation rate by subtype. Head Neck. 2020;42:539–555. doi: 10.1002/hed.26006. [DOI] [PubMed] [Google Scholar]
- 54.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat. Rev. Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 55.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willis O, et al. PIK3CA gene aberrancy and role in targeted therapy of solid malignancies. Cancer Gene Ther. 2020;27:634–644. doi: 10.1038/s41417-020-0164-0. [DOI] [PubMed] [Google Scholar]
- 58.Sharma SV, Settleman J. Oncogene addiction: setting the stage for molecularly targeted cancer therapy. Genes Dev. 2007;21:3214–3231. doi: 10.1101/gad.1609907. [DOI] [PubMed] [Google Scholar]
- 59.Moody CA, Laimins LA. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog. 2009;5:e1000605. doi: 10.1371/journal.ppat.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Albert, E. & Laimins, L. Regulation of the human papillomavirus life cycle by DNA damage repair pathways and epigenetic factors. Viruses12, 744 (2020). [DOI] [PMC free article] [PubMed]
- 61.Shibata, A. & Jeggo, P. A. ATM’s role in the repair of DNA double-strand breaks. Genes12, 1370 (2021). [DOI] [PMC free article] [PubMed]
- 62.Shaikh MH, et al. Chromosome 3p loss in the progression and prognosis of head and neck cancer. Oral Oncol. 2020;109:104944. doi: 10.1016/j.oraloncology.2020.104944. [DOI] [PubMed] [Google Scholar]
- 63.Dasari A, Grothey A, Kopetz S. Circulating tumor DNA-defined minimal residual disease in solid tumors: opportunities to accelerate the development of adjuvant therapies. J. Clin. Oncol. 2018;36:3437–3440. doi: 10.1200/JCO.2018.78.9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim S, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods. 2018;15:591–594. doi: 10.1038/s41592-018-0051-x. [DOI] [PubMed] [Google Scholar]
- 66.Taylor-Weiner A, et al. DeTiN: overcoming tumor-in-normal contamination. Nat. Methods. 2018;15:531–534. doi: 10.1038/s41592-018-0036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cibulskis K, et al. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics. 2011;27:2601–2602. doi: 10.1093/bioinformatics/btr446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taylor-Weiner A, et al. Scaling computational genomics to millions of individuals with GPUs. Genome Biol. 2019;20:228. doi: 10.1186/s13059-019-1836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim J, et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat. Genet. 2016;48:600–606. doi: 10.1038/ng.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Therneau, T. M. A package for survival analysis in R. https://CRAN.R-project.org/package=survival (2021).
- 72.Harrell, F. E. Jr rms: regression modeling strategies. https://CRAN.R-project.org/package=rms (2021).
- 73.R Core Team R: a language and environment for statistical computing (R Foundation for Statistical Computing, 2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Event prevalence comparison between HPV+ and HPV– HNSCC. a, Number and percentage of cases having each of 66 genomic events for 421 HPV– and 101 HPV+ HNSCCs. Fisher’s exact test two-sided P values are reported for comparison between HPV– and HPV+ tumors. Benjamini–Hochberg false discovery rate-corrected q values are also shown. b, List of differentially prevalent events at q < 0.1. c, List of events not differentially prevalent (q < 0.1). d, Differentially prevalent events in b favored in HPV– HNSCC. e, Differentially prevalent events in b favored in HPV+ HNSCC. Supplementary Table 2. Clonal event prevalence comparison between HPV+ and HPV– HNSCC, like Supplementary Table 1 but restricted to clonal events. Supplementary Table 3. Event prevalence by anatomic subsite for HPV– HNSCC. Breakdown by anatomic subsite of number and percentage of cases having specified genomic events: larynx, 114 tumors; oral cavity, 274 tumors; oropharynx, 33 tumors. Fisher’s exact test P values and Benjamini–Hochberg q values are as in Supplementary Table 1. Supplementary Table 4. Event prevalence comparison between NSD1-mutated and other HPV– HNSCC, like Supplementary Table 1a–c but comparing 54 NSD1-mutated HPV– HNSCC against 367 other HPV– HNSCC. Supplementary Table 5. Event prevalence comparison between CASP8-mutated and other HPV– HNSCC, like Supplementary Table 1a–c but comparing 50 CASP8-mutated HPV– HNSCCs against 371 other HPV– HNSCCs. Supplementary Table 6. Event prevalence comparison between NOTCH1-mutated and other HPV– HNSCCs, like Supplementary Table 1a–c but comparing 70 NOTCH1-mutated HPV– HNSCCs against 342 other HPV– HNSCCs. Supplementary Table 7. Real-time timing estimates for whole-genome copy-number events.
Data Availability Statement
New WES data that support the findings of this study have been deposited in dbGaP under accession code phs003139.v1. Other human HNSCC genomic data were derived from the TCGA Research Network at http://cancergenome.nih.gov/. Source data are provided with this paper. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
The PhylogicNDT package is available for download from https://github.com/broadinstitute/PhylogicNDT.