Extended Data Fig. 16. Astrocyte subcompartment card 6: astrocyte gap junction protein.
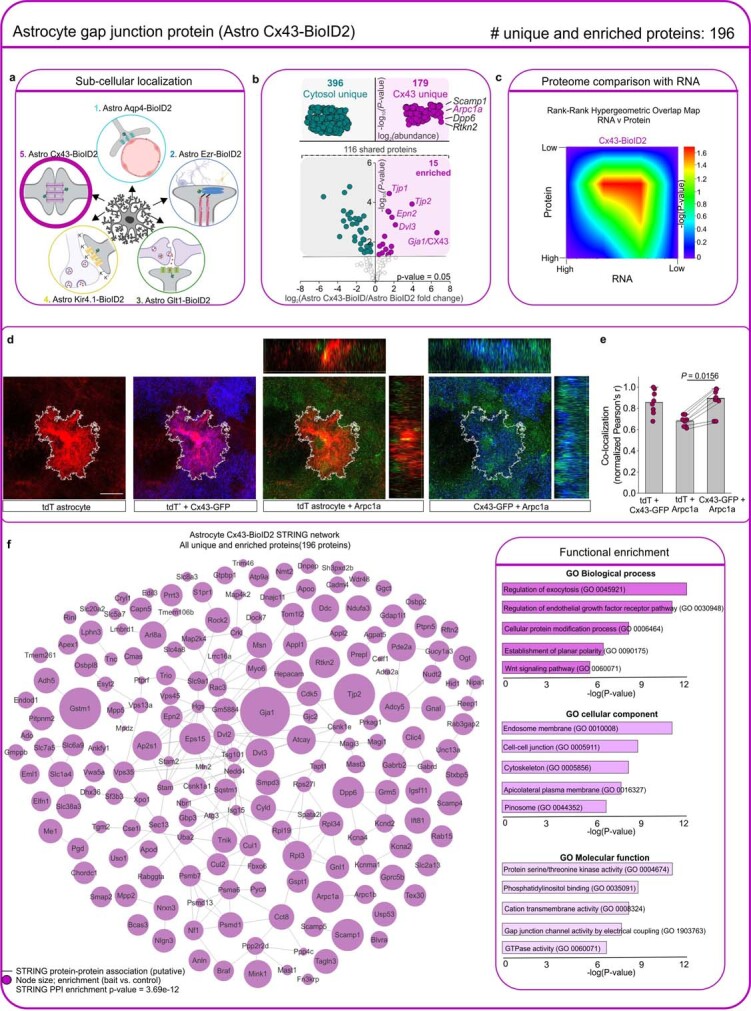
a. BioID2 that is targeted with Cx43-BioID2 biotinylates proteins at astrocyte sites of gap junction coupling. b. Label-free based quantification comparison of significant proteins (Log2FC > 1 and FDR < 0.05 versus GFP controls) detected in the cytosolic Astro BioID2 and Astro Cx43-BioID2 reveal Cx43 enriched proteins. Top half of the volcano plot shows 179 unique Cx43-BioID2 proteins when compared to cytosol. The top four most abundant proteins for Cx43-BioID2 are shown. Lower half of volcano plot shows comparison of 116 proteins that were common in both cytosolic BioID2 and Cx43-BioID2. The five highest enriched proteins for Cx43-BioID2 are shown. Magenta label shows protein that was validated with immunohistochemistry. c. Heat map shows the rank-rank hypergeometric overlap (RRHO) of the RNA and protein rank for the 196 Cx43-BioID2 proteins. Each pixel represents the significance of overlap between the two datasets in –log10(P-value). Red pixels represent highly significant overlap. Color scale denotes the range of P-values at the negative log10 scale (bin size = 100). d. IHC analysis of Arpc1a protein in tdTomato and Cx43-GFP labeled astrocytes shows co-localization within the astrocyte territory. Scale bar represents 20 μm. e. Co-localization analysis using Pearson’s r co-efficient shows high co-localization between Cx43-GFP and Aprc1a. The mean and SEM are shown (n = 8 tdTomato+ cells from 4 mice; Two-tailed Wilcoxon matched-pairs signed rank test). f. Scale-free STRING analysis protein-protein association map of the 196 unique and enriched biotinylated proteins identified with Astro Cx43-BioID2 . Node size represents the enrichment of each protein vs the GFP control (log2(BioID2/GFP)). Edges represent putative interactions from the STRING database. Bar graphs show the functional enrichment analysis of all 196 proteins using “Biological process”, “Cellular component”, and “Molecular function” terms from Enrichr. The image of the astrocyte subcompartments in panel a was created using BioRender (https://www.biorender.com/).
