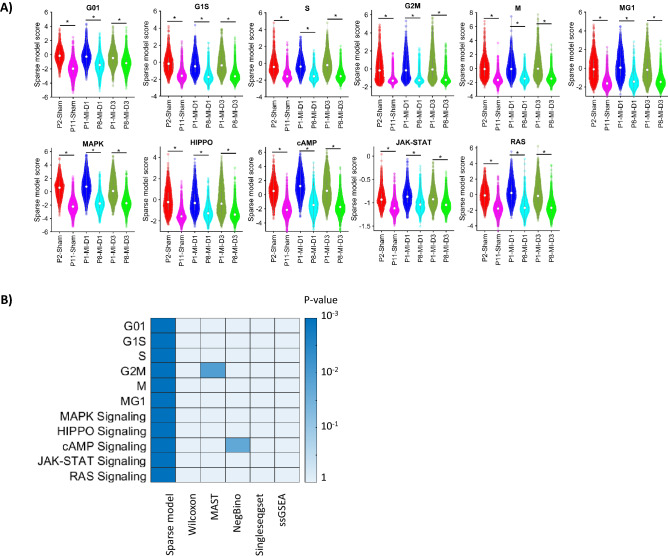
Figure 4.
AI Sparse Modeling was more effective than non-AI tools for pathway/gene set enrichment analysis of cardiomyocyte scRNAseq data from mouse hearts. (A) Sparse model scores for each cell-cycle phase and for the activity of the MAPK, HIPPO, cAMP, JAK-STAT, and RAS signaling pathways were summarized for cardiomyocytes from the indicated injury groups and time points and presented as violin plots. Statistical comparison between the P1-MI-D1/D3 (regenerative) and P8-MI-D1/D3 (non-regenerative), also between P2-Sham and P11-Sham, were done by non-parametric tests; *p-value < 0.01. (B) Cardiomyocytes from the P1-MI-D3 and P8-MI-D3 groups were scored for cell-cycle phase and pathway activity via AI Sparse Modeling, Wilcoxon Ranksum test, MAST, Negative Binomial (NegBino) test, Singleseqset, and ssGSEAP; then, the scores generated by each technique were compared between time points, and the P-values for each comparison was presented as a heat-map.